Project 2: Introduction to the background and theory
BioSS/UKCEH
BioSS
Problem specification
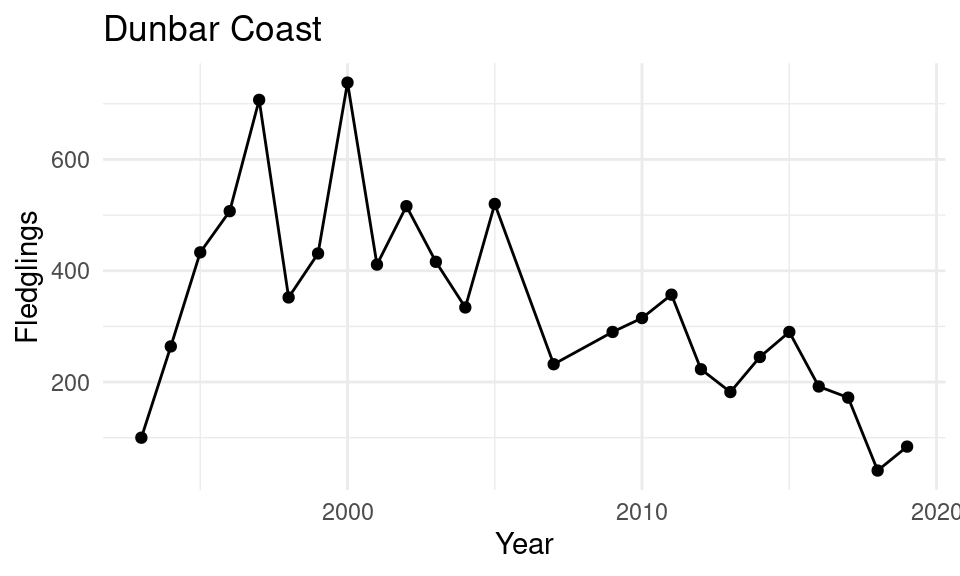
Kittiwakes: how are they doing?
- Measure: “breeding success”
- Number of fledglings per year
- (From “apparently occupied nests”)
A basic model
Breeding success is a function of time
\[ \texttt{Fledg}_t = \exp\left(\beta_0 + \texttt{AON}_t + s(t)\right) \]
where \(\texttt{Fledg}_t \sim \text{Tweedie}(\phi, p)\).
- \(t\) indexes years
- \(\beta_0\) is an intercept
- \(\texttt{Fledg}_t\) is the number of fledglings
- \(\texttt{AON}_t\) number of apparently occupied nests
- \(s(t)\) smooth of year
In R…
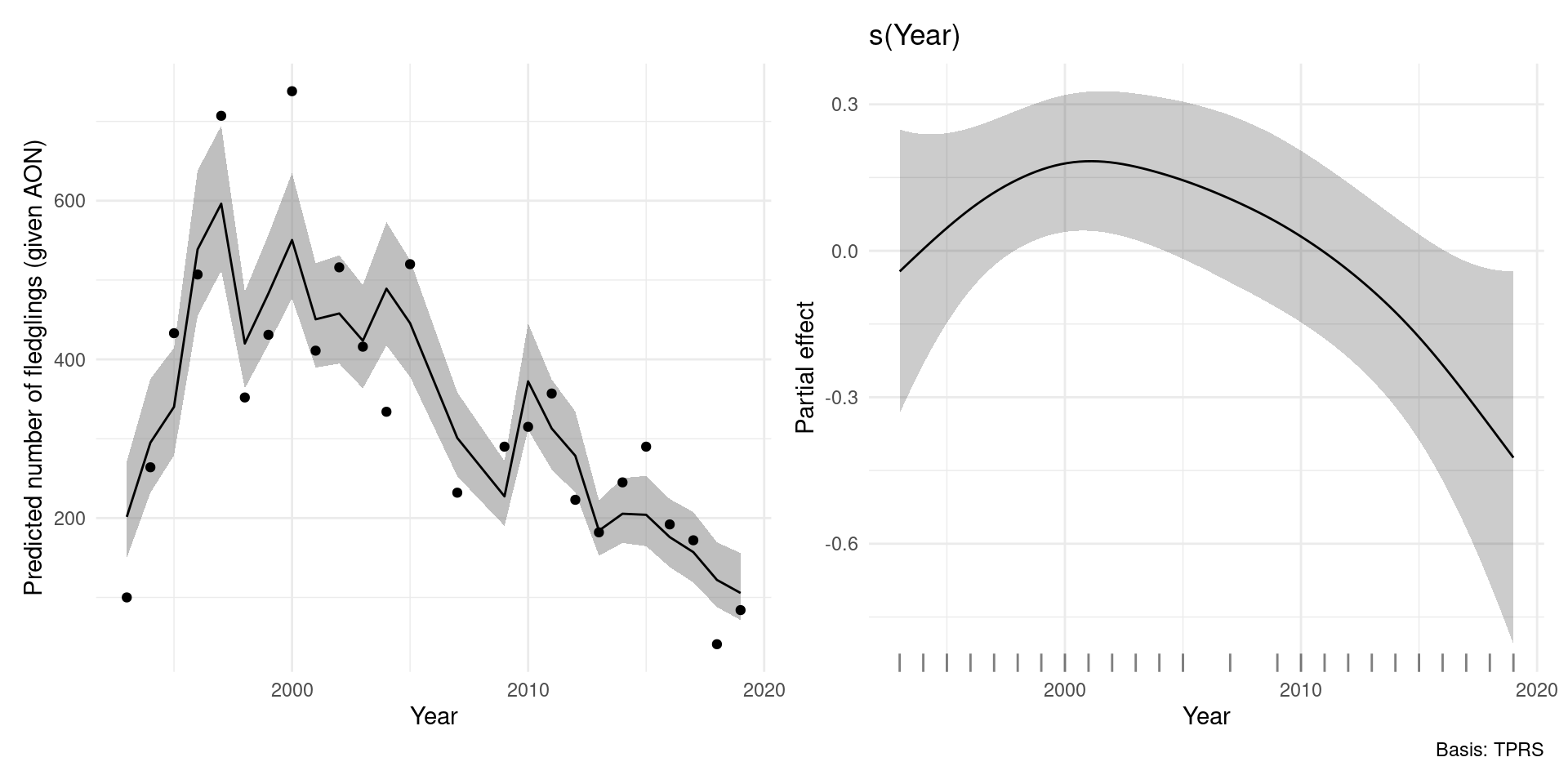
That’s all well and good, but…
We need to do some ecology?
- We want to know why
- What influences this?
- (Remotely sensed) covariates?
- sea surface temperature
- lesser sandeel Ammodytes marinus distribution
- (Remotely sensed) covariates?

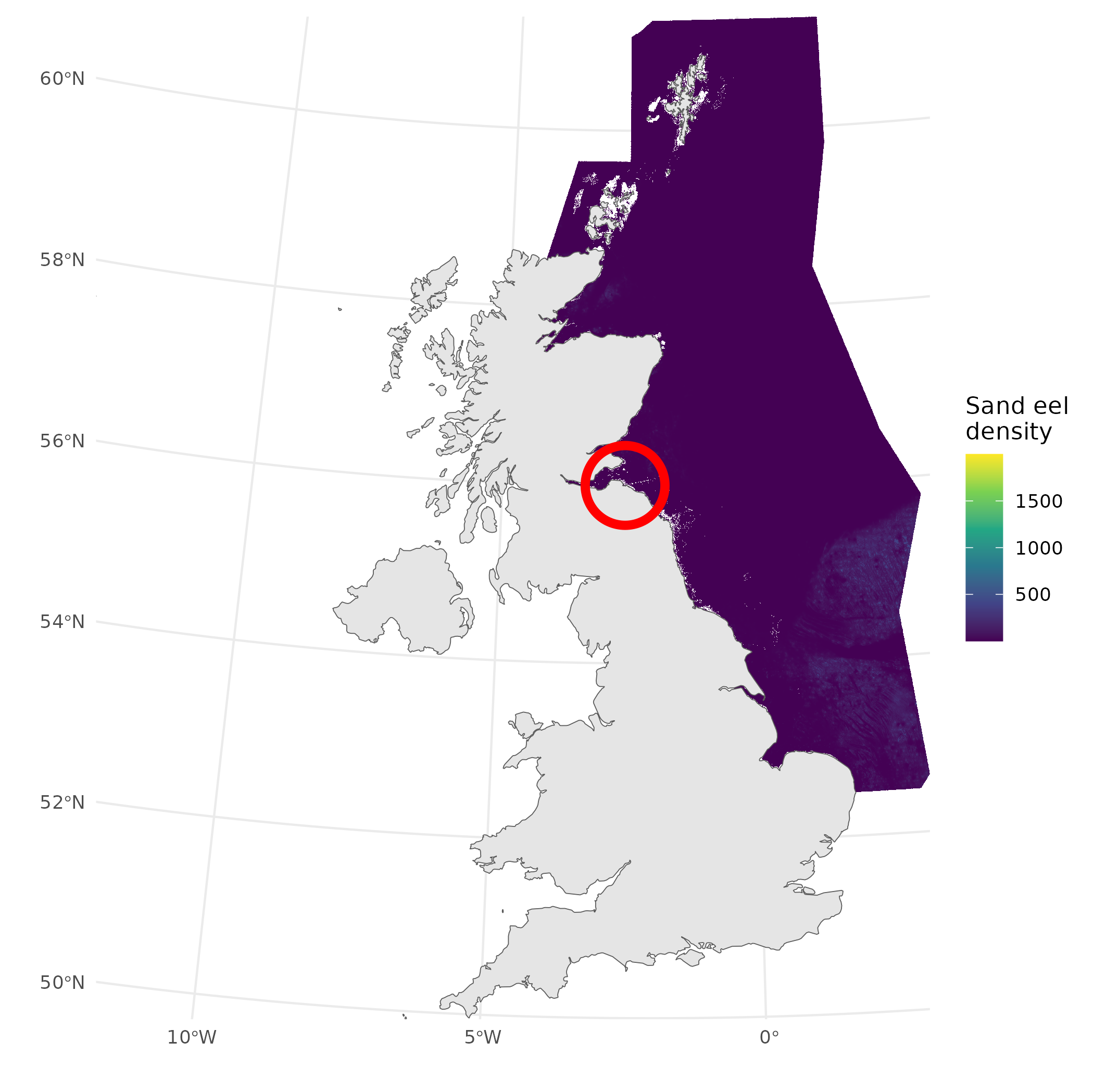
Sand eels
- We have a map of sandeel distribution
- We can just “plug-in” values, right?
Langton, R., Boulcott, P. and Wright P.J. (2021) A verified distribution model for the lesser sandeel Ammodytes marinus. Marine Ecology Progress Series.

What does that look like?
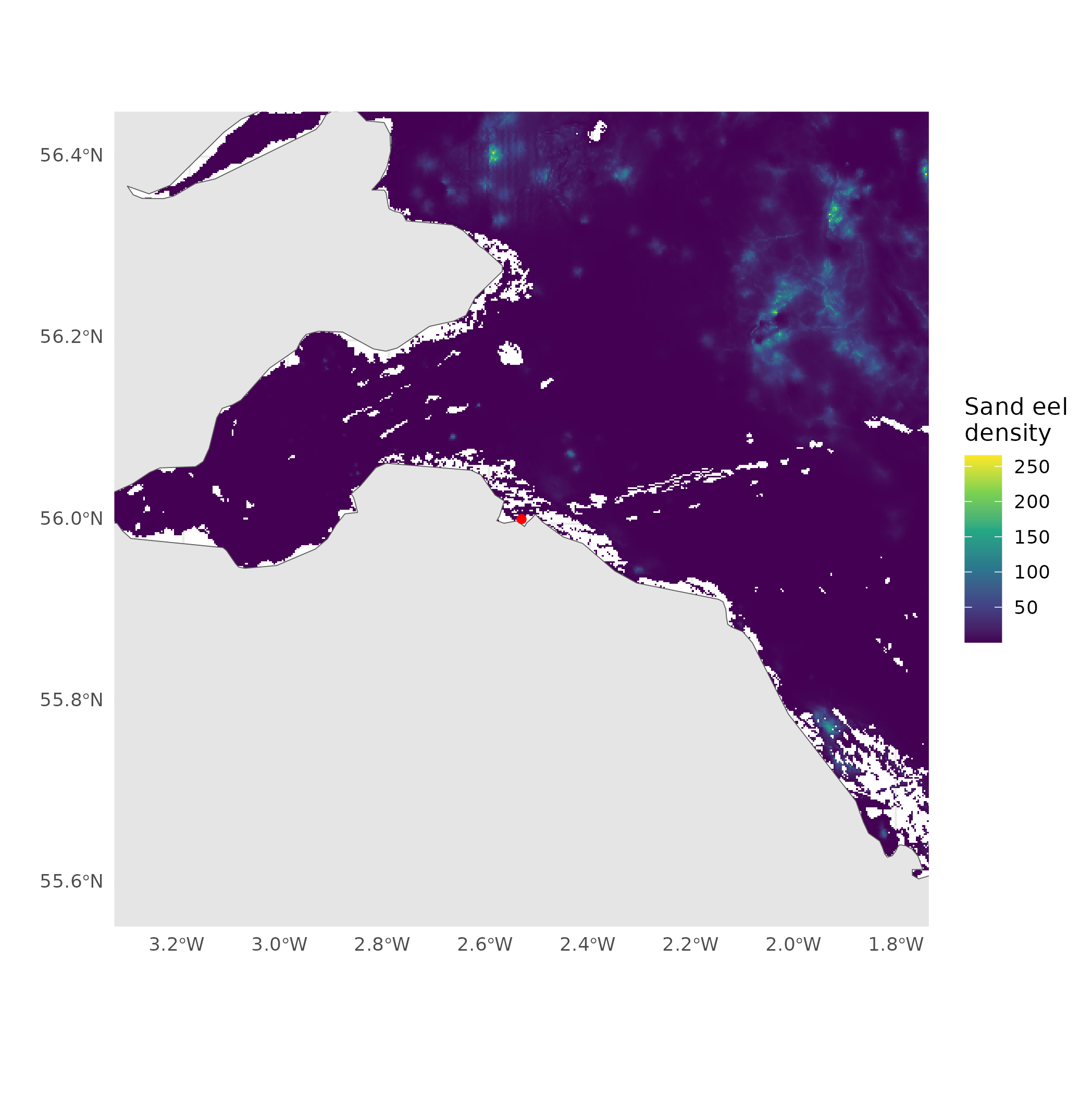
(zoom and enhance)
- Which value should we take?
- Closest?
- Average?
- At what distance?
- What does the literature say?
Let’s simplify that
- We know a little about foraging distance
- Rings (annuli 🧐)
- Simplifying assumption
- 10km wide, up to 100km away
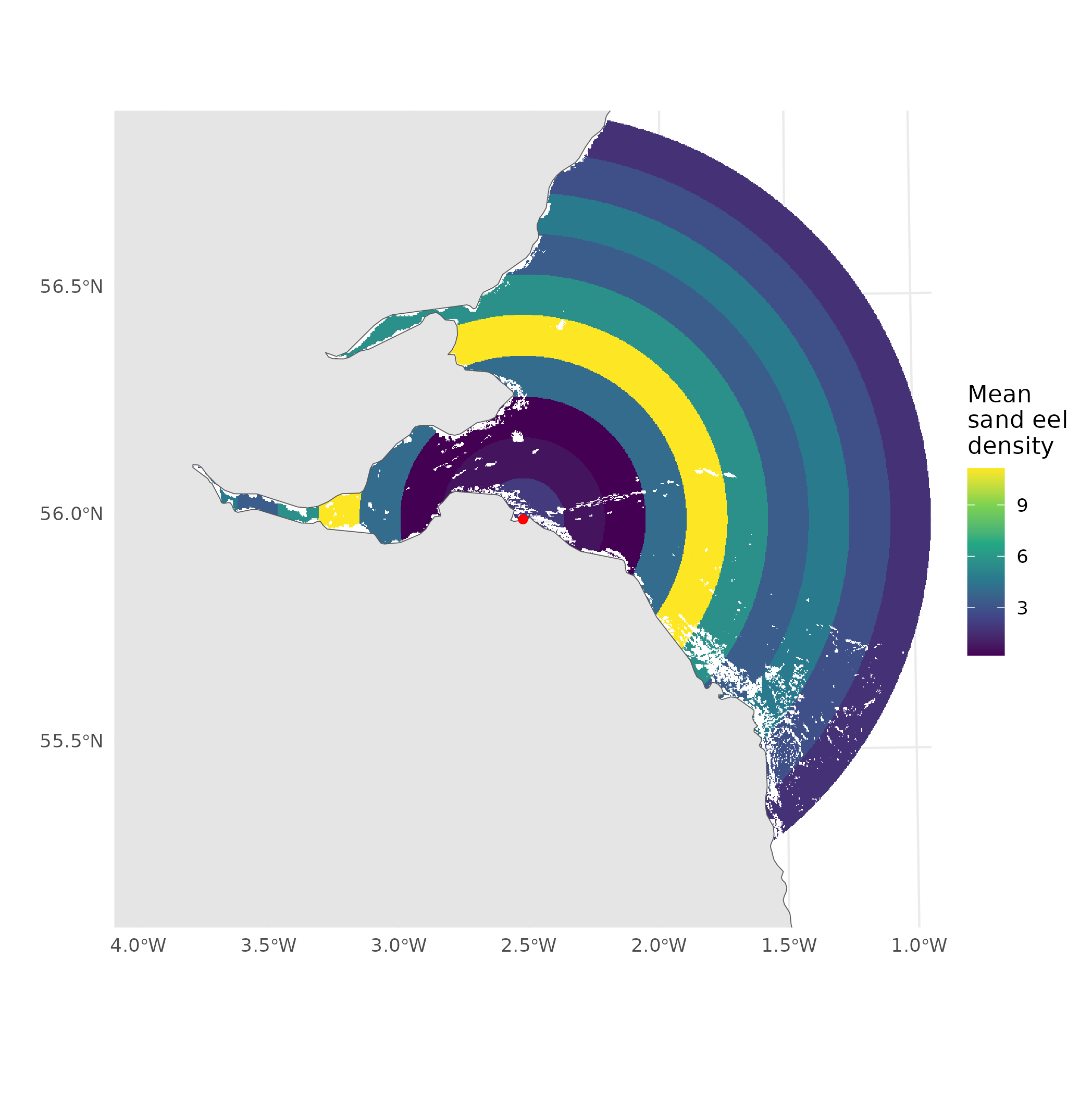
How can we build a model for this?
New problem specification
- We now have 10 values (10 rings) of sandeel density
- We don’t know which one is “right”
- 👻 maybe more than one is right 👻
- How can we include 10 values in the model?

Approaches from literature
- Put all the covariates in the model
eel1 + eel2 + ...- Colinearity?
- Independence??
- Fit a LOT of models
- Which is best (by AIC?)?
- Is this a good idea? (No)

🧠 Extending to multiple sites
- Can we learn something about general kittiwake ecology?
- What can we generalize?
- What can we say about overall trends?
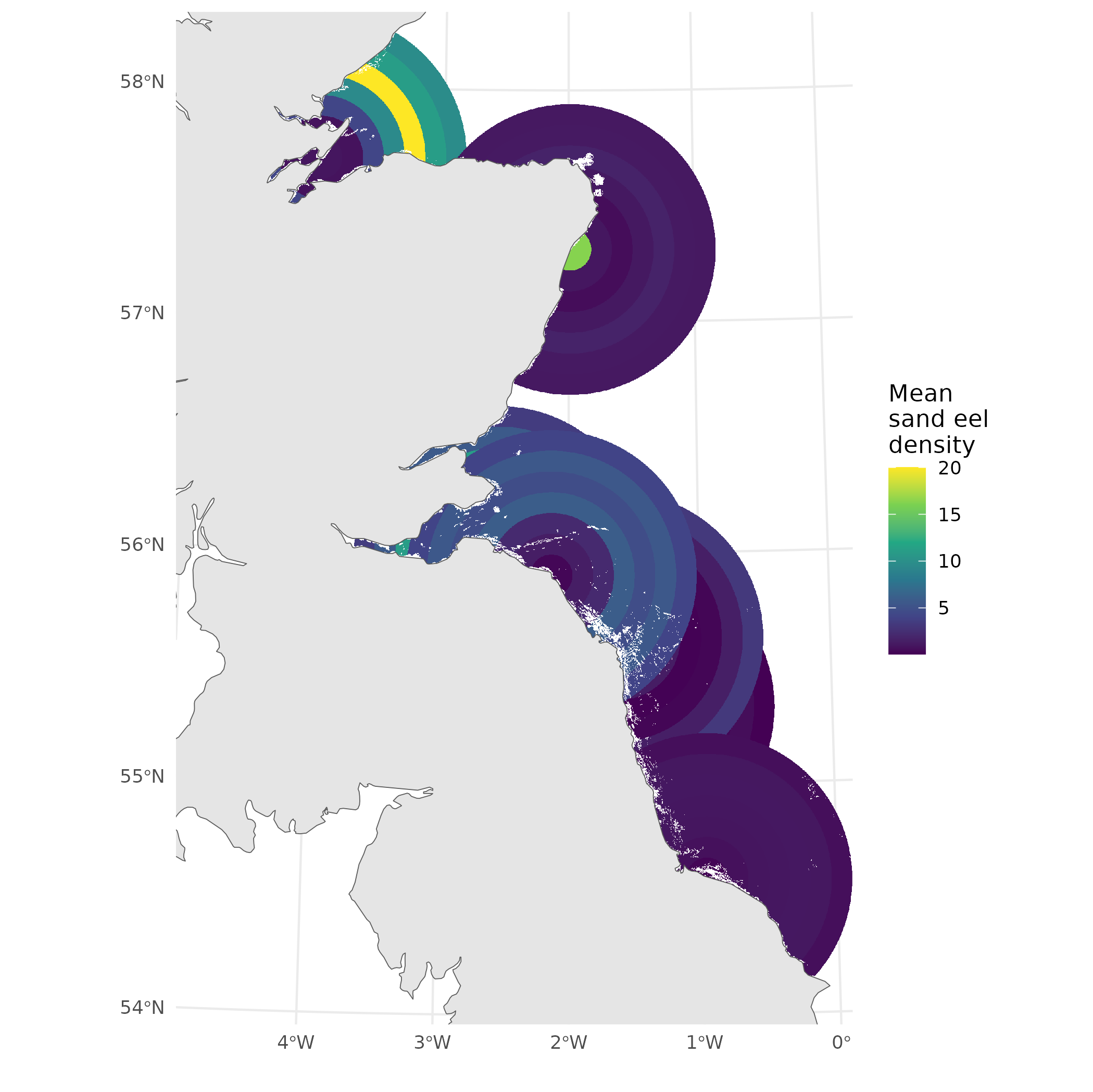
What do we really want here?
- Some average over the rings, but how do we weight them?
- What if we could weight while fitting?
- more efficient use of information
- (hint: that’s what we’ll do)

What does that data actually look like?
- (Restricting to 80km radius)
- We have 8 data points
- For one site
- Can we get the model to select which one(s) are useful?
- (Yes)
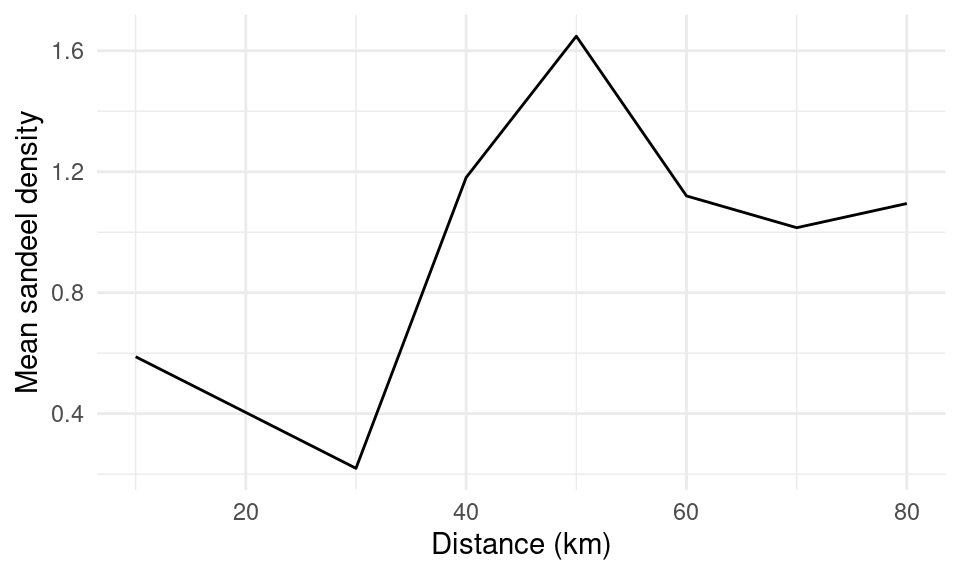
Weighted averaging
- To estimate a weighted mean:
\[ \sum_j w_j x_j \]
- what if \(w_j\) and \(x_j\) are functions?
\[ \sum_j w(z_j) x(z_j) \]
Weighted averaging in a GAM
\[ \sum_j w(z_j) x(z_j) \]
\(w(z_j)\) is a smooth we estimate
\(x(z_j)\) is the sandeel density
\(z_j\) are the distances
\(\Rightarrow\) weighted average of sandeel density over the distances
- where we estimate the weighting
Let’s do that…
Enter signal regression
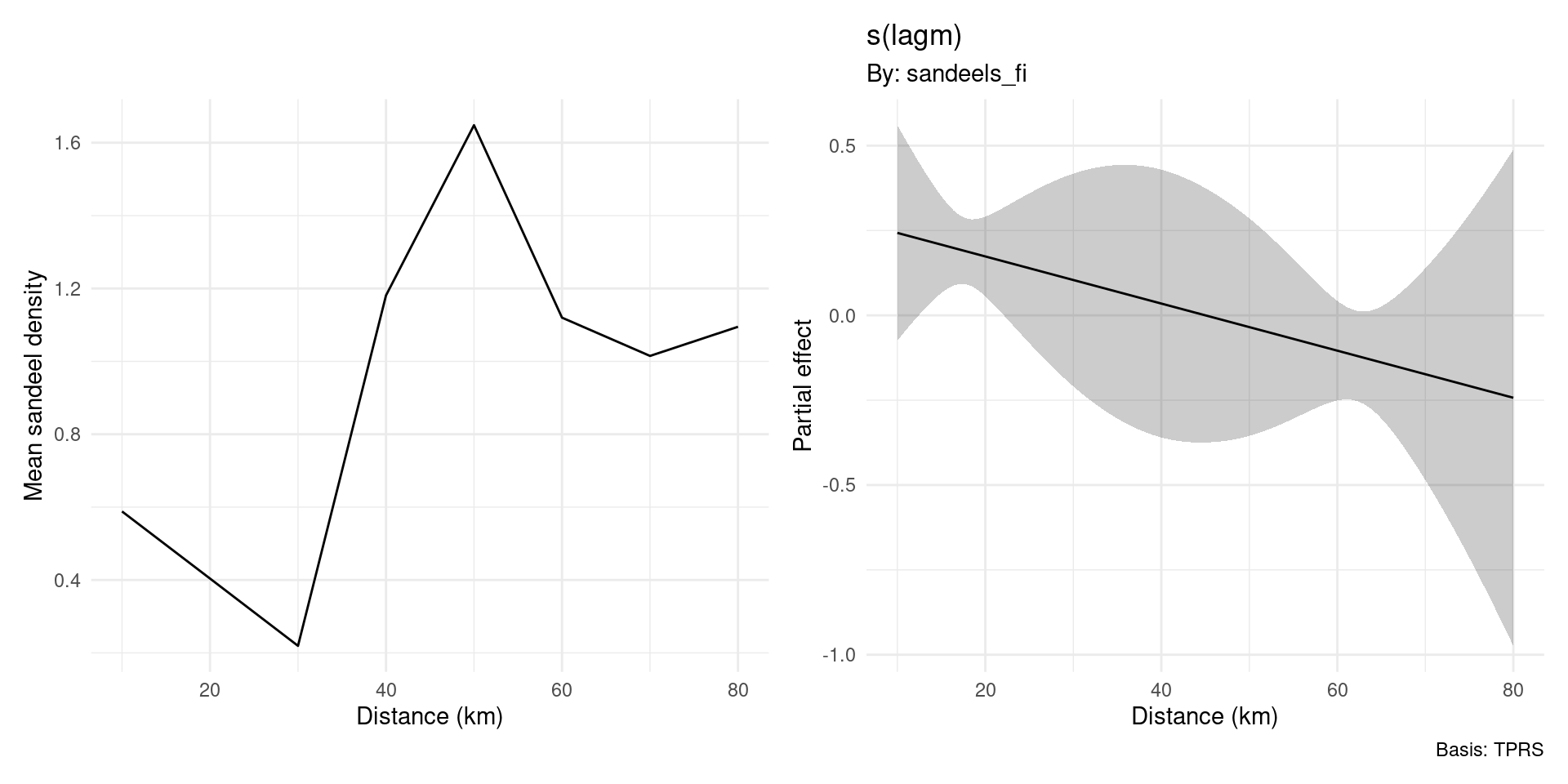
Signal regression, in maths
\[ \texttt{Fledg}_t = \exp\left( \beta_0 + \texttt{AON}_t + s(t) + \sum_j w(d_j) e(d_j) \right) \]
- Everything else as before but now an extra weighted sum
- \(d_j\) are the distance “lags” we have sandeel density at
- \(w(d_j)\) is our smooth of distance
- \(e(d_j)\) is the value of sand eel density at distance \(d_j\)
Signal regression, in R
“estimate a smooth (the weighting) of lagm (distances) and multiply by the sandeel density at that distance”
What does that data look like?
lagmandsandeels_fiarenrow(kit_sub)\(\times\) (number of distances) matrices
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8]
[1,] 10 20 30 40 50 60 70 80
[2,] 10 20 30 40 50 60 70 80
[3,] 10 20 30 40 50 60 70 80
[4,] 10 20 30 40 50 60 70 80
[5,] 10 20 30 40 50 60 70 80
[6,] 10 20 30 40 50 60 70 80 [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8]
[1,] 0.5881714 0.4039171 0.21924 1.180914 1.648092 1.119965 1.01492 1.094924
[2,] 0.5881714 0.4039171 0.21924 1.180914 1.648092 1.119965 1.01492 1.094924
[3,] 0.5881714 0.4039171 0.21924 1.180914 1.648092 1.119965 1.01492 1.094924
[4,] 0.5881714 0.4039171 0.21924 1.180914 1.648092 1.119965 1.01492 1.094924
[5,] 0.5881714 0.4039171 0.21924 1.180914 1.648092 1.119965 1.01492 1.094924
[6,] 0.5881714 0.4039171 0.21924 1.180914 1.648092 1.119965 1.01492 1.094924Recap
We can use signal regression (aka scalar-on-function regression) to make weighted averages of “functional” data.
We assume that sandeel density is a function of distance from colony.
We estimate the weighting needed during fitting.
Adding complication: more sites
Adding sites
- Want to look at colony status over multiple sites
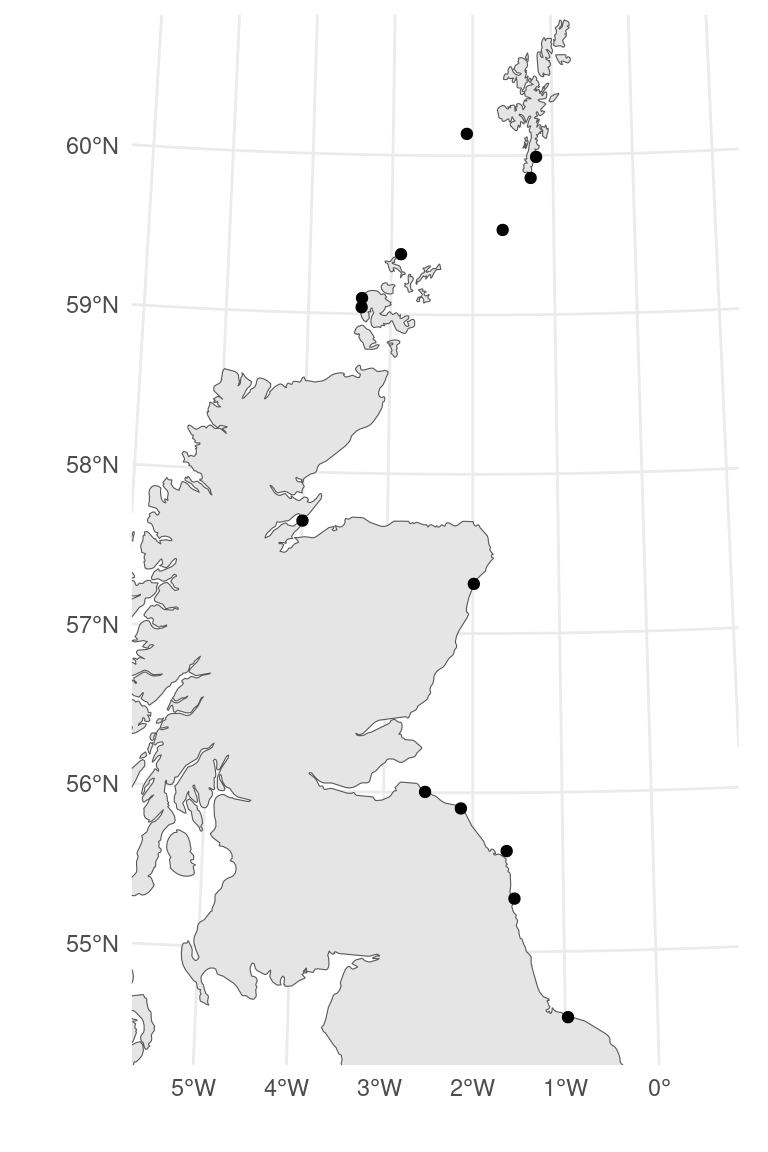
- Let’s include data from 14 sites
- (where we have the sandeel data)
Adding site-specific effects (maths)
\[\begin{equation} \texttt{Fledg}_{ti} = \exp\left( \beta_0 + \texttt{AON}_{ti} + s(t) +\\ \qquad \sum_j w(d_j) e_i(d_j) + \texttt{Site}_i \right) \end{equation}\]
- (extra index \(i\) for site)
- AON is now site (\(i\)) and time (\(t\)) specific
- site random intercept \(\texttt{Site}_i\)
- sandeel density per site \(e_i()\)
- but still looking for common distance smooth!
Adding site-specific effects (R)
What does that look like?
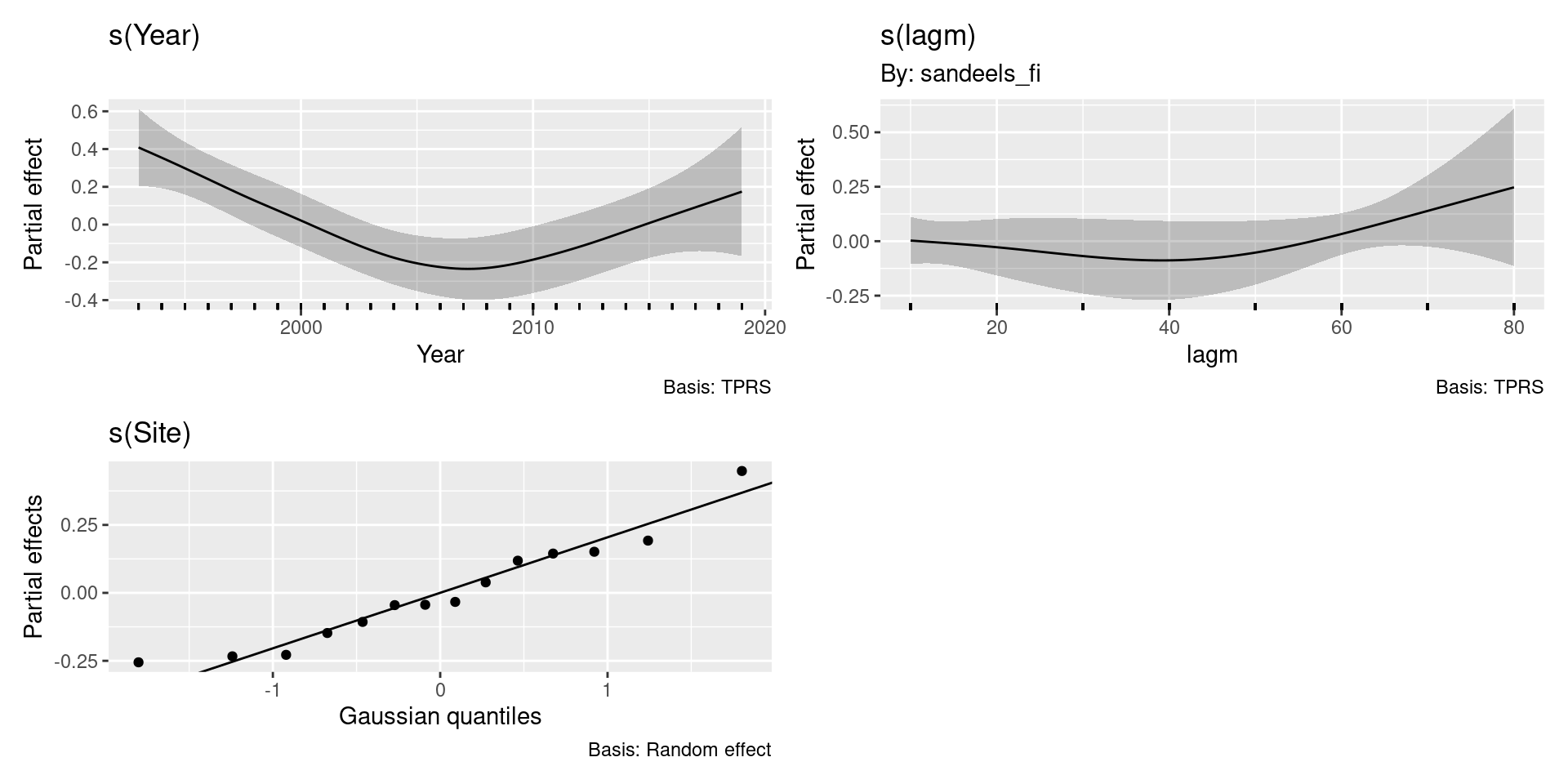
Some things to think about here
- How reasonable is this model?
- Do we expect that the distance effect is the same for all sites?
- Should we have a spatial effect rather than site random effect?
- Are there other plausible models?
But we need to keep going…
More complicated example using SST
Sea surface temperature
- How should we include SST in the model?
- Expand our model like this?
\[ \texttt{Fledg}_t = \exp\left(\beta_0 + \texttt{AON}_t + s(t) + s(\texttt{SST}_t)\right) \]
- Easy, right?
Things are never easy…
- We have weekly SST
- We have SST on a 10km grid
- Which one is important?
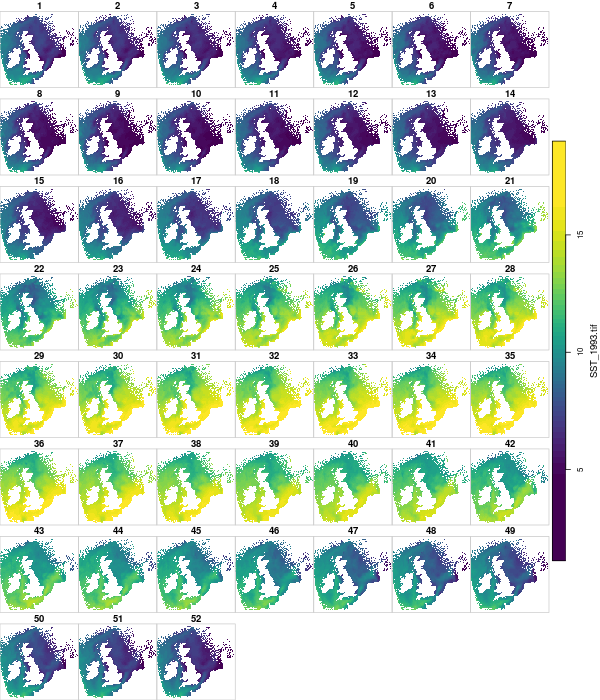
Going back to just Dunbar
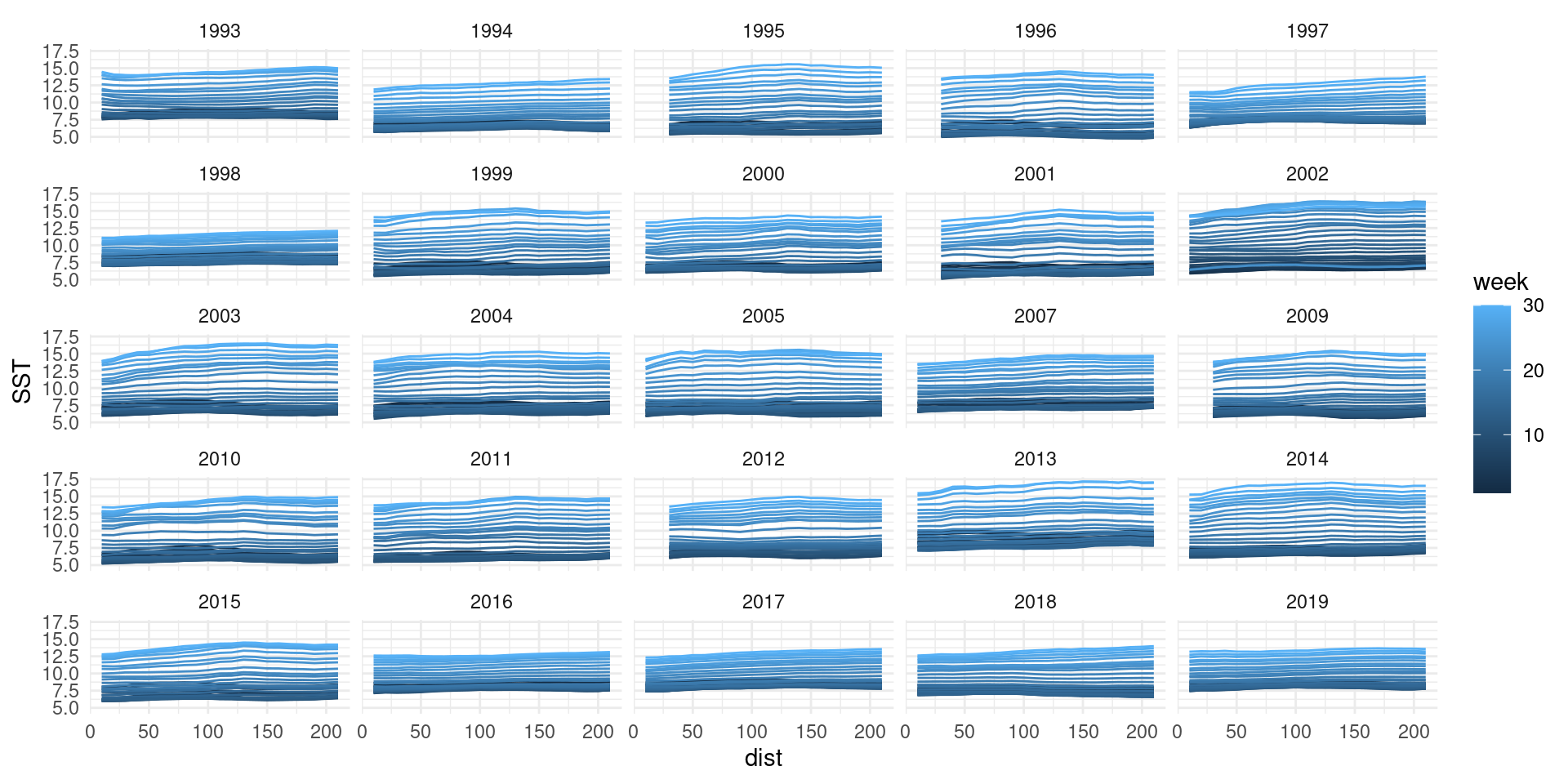
That’s a lot
We can do the same trick though!
- Let’s simplify again using rings
- Two lags: time and distance
- Let’s do all sites
- and let the smooth vary by site
What can we get to?
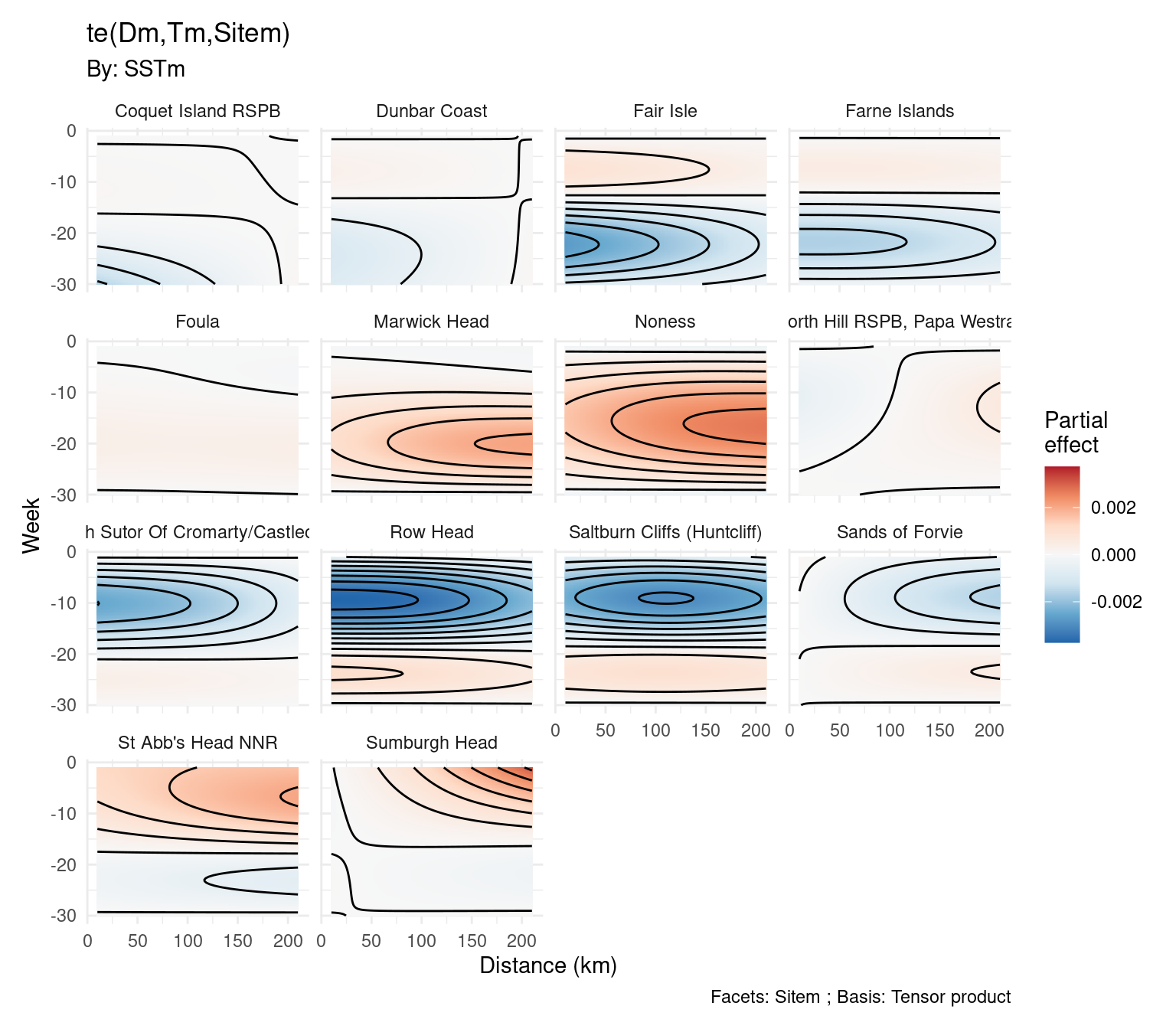
What just happened?
No, really
- We made another average but it’s really fancy now
- For a given \(\texttt{Site}\)
\[ \sum_j \sum_k w(d_j, t_k, \texttt{Site}) \texttt{SST}(d_j, t_k, \texttt{Site}) \]
- \(w(d_j, t_k, \texttt{Site})\) is a smooth we estimate
- \(\texttt{SST}(d_j, t_k, \texttt{Site})\) is the SST
- \(d_j\) are the distances
- \(t_k\) is the week
How do we build \(w(d_j, t_k, \texttt{Site})\)
- Let’s start with just making a smooth of 2 variables
- \(s(x, y)\)
- but \(x\) and \(y\) are on different scales
- Tensor products!
Tensor what?
Looking for a smooth function \(f(X1, X2)\)
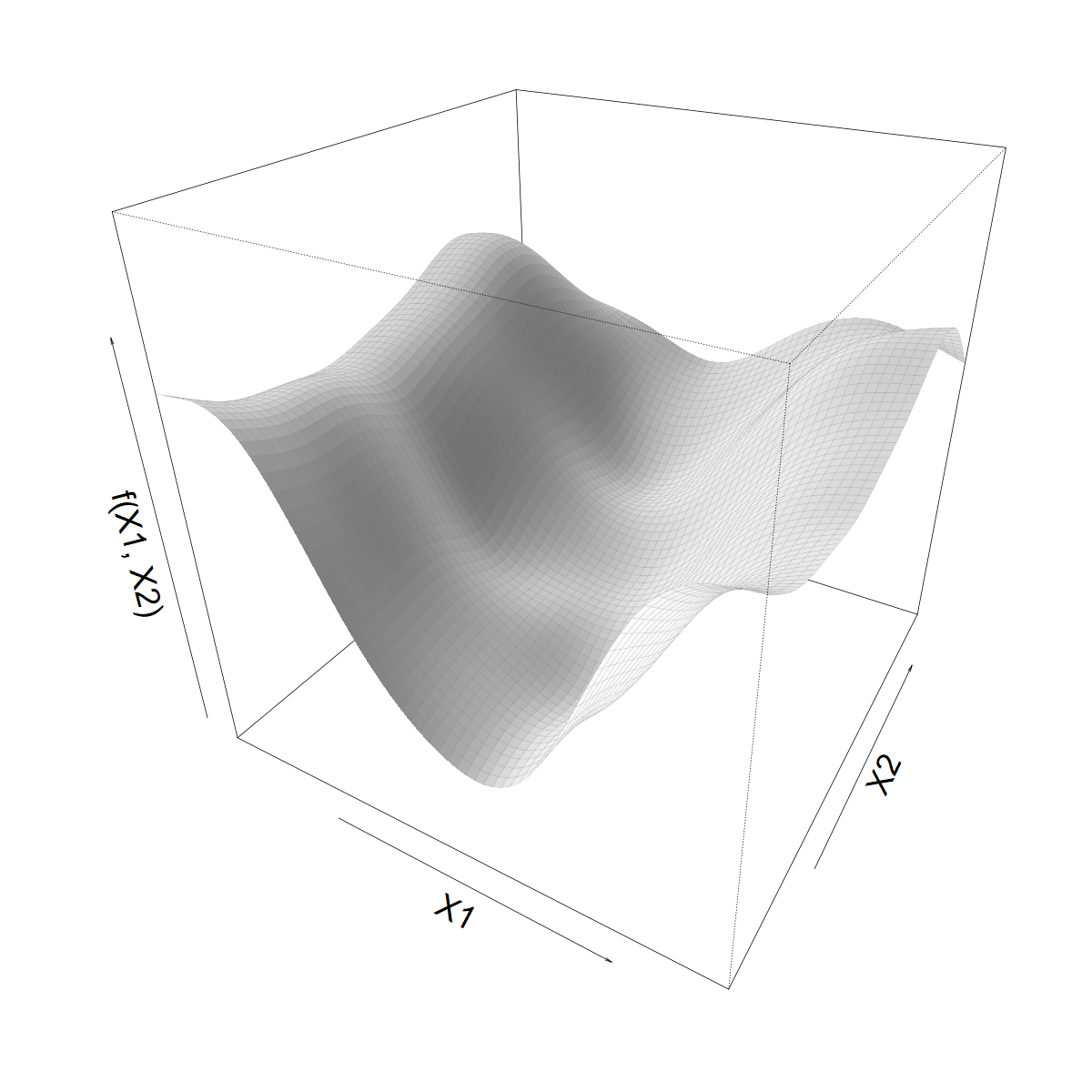
Smoothing over \(X1\)
- Let’s start with a standard 1D smoother
\[ s(X1) = \sum_k \beta_k b_k(X1) \]
- Built from basis functions \(b_k(X1)\)
- With coefficients \(\beta_k\) (“weights”)

Smoothing over \(X1\)
- Let’s start with a standard 1D smoother
\[ s(X1) = \sum_k \beta_k b_k(X1) \]
Built from basis functions \(b_k(X1)\)
With coefficients \(\beta_k\) (“weights”)
We want to let \(s(X1)\) vary with \(X2\)
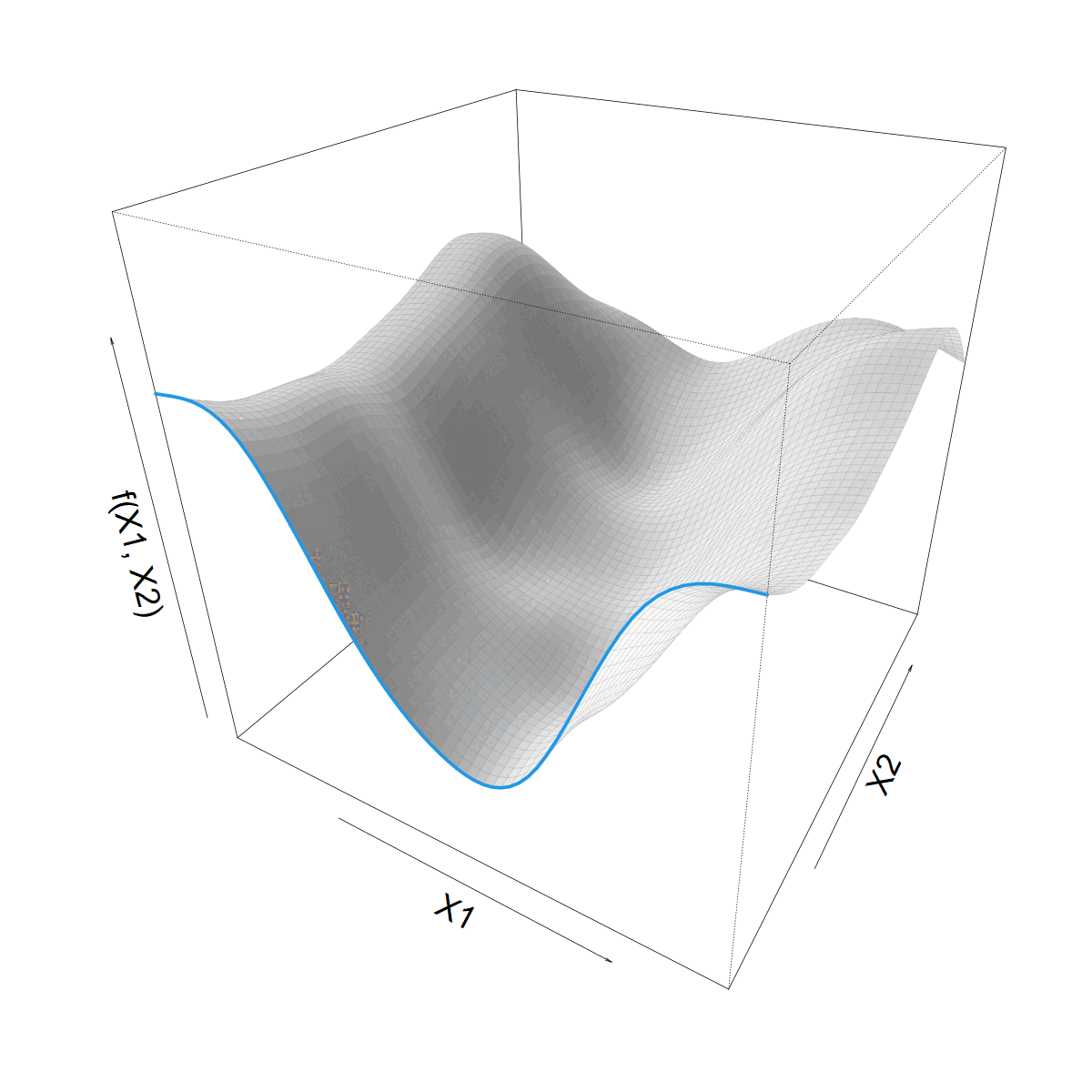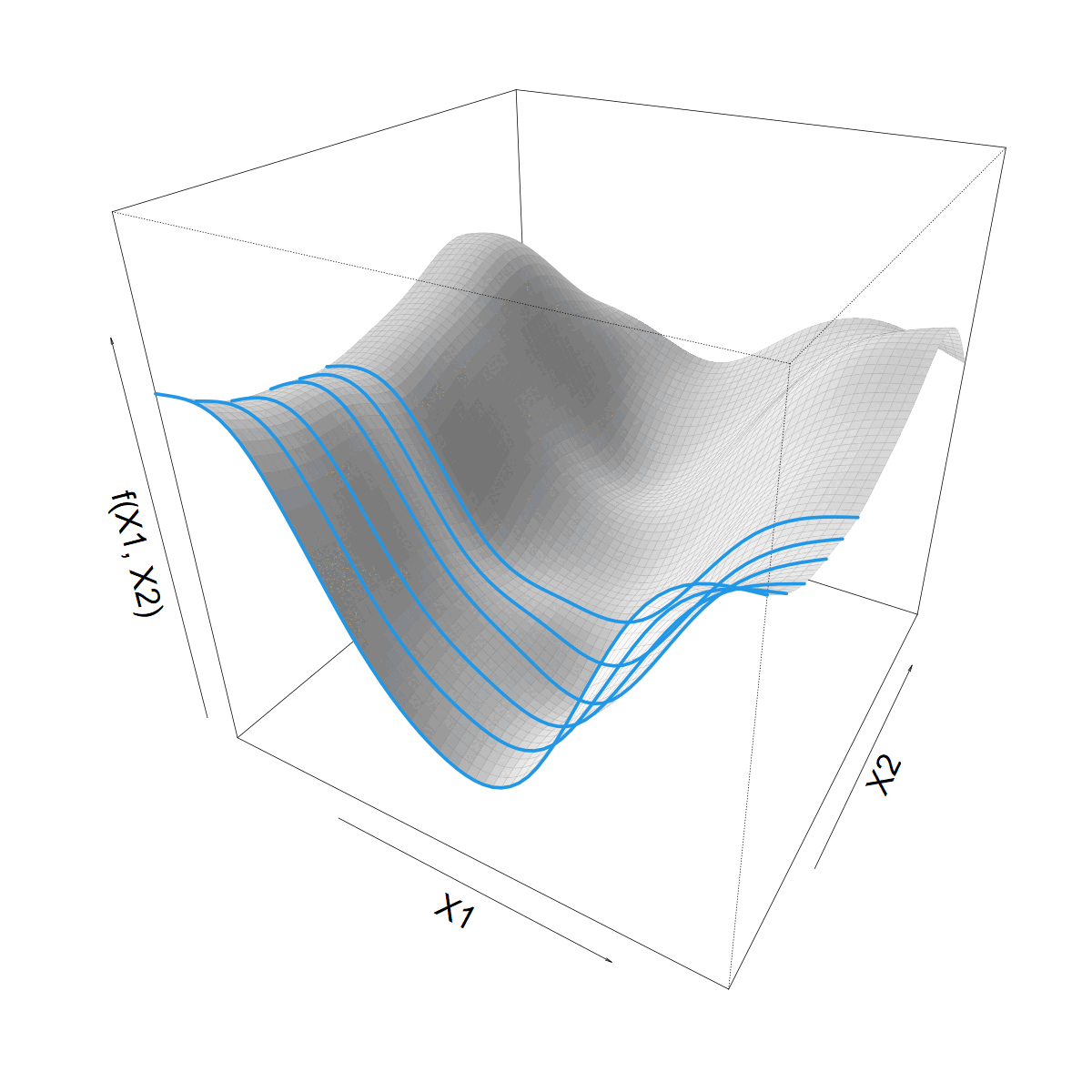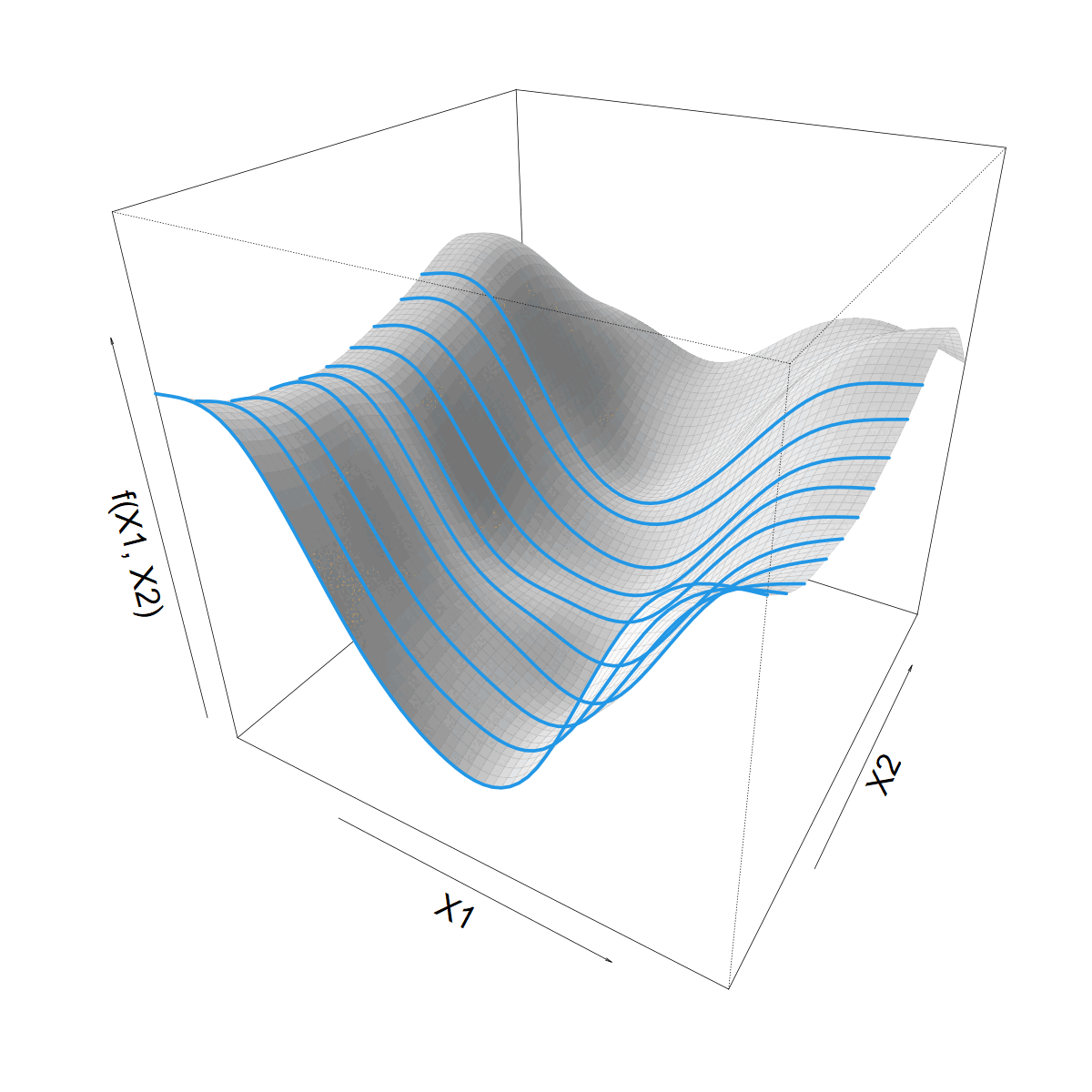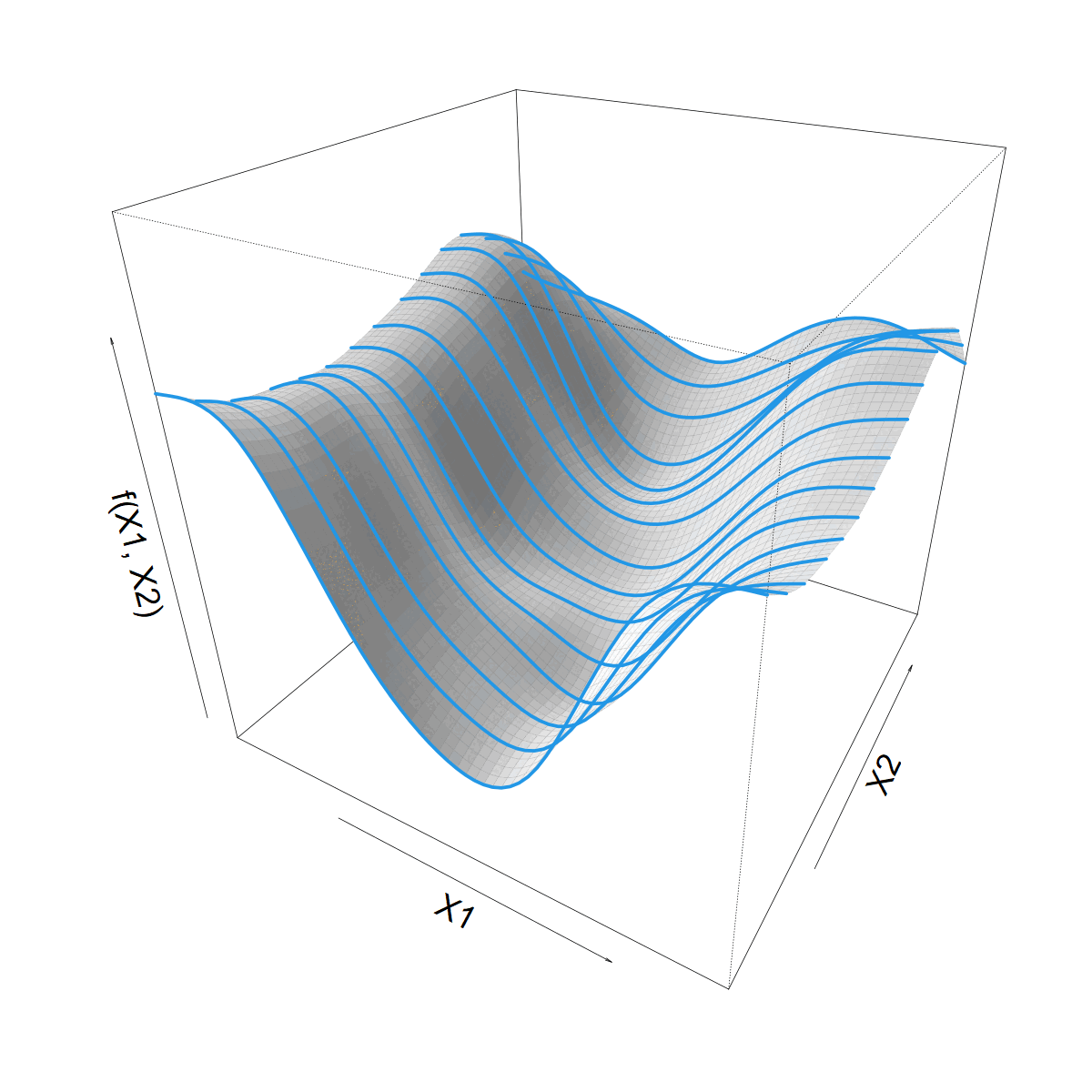
Making \(s(X1)\) vary smoothly with \(X2\)
\[ s(X1) = \sum_k \beta_k b_k(X1) \]
\(\beta_k\) now 1D smooth function of \(X2\)
\[ \beta_k(X2) = \sum_j \alpha_{jk} a_j(X2) \]
Making \(s(X1)\) vary smoothly with \(X2\)
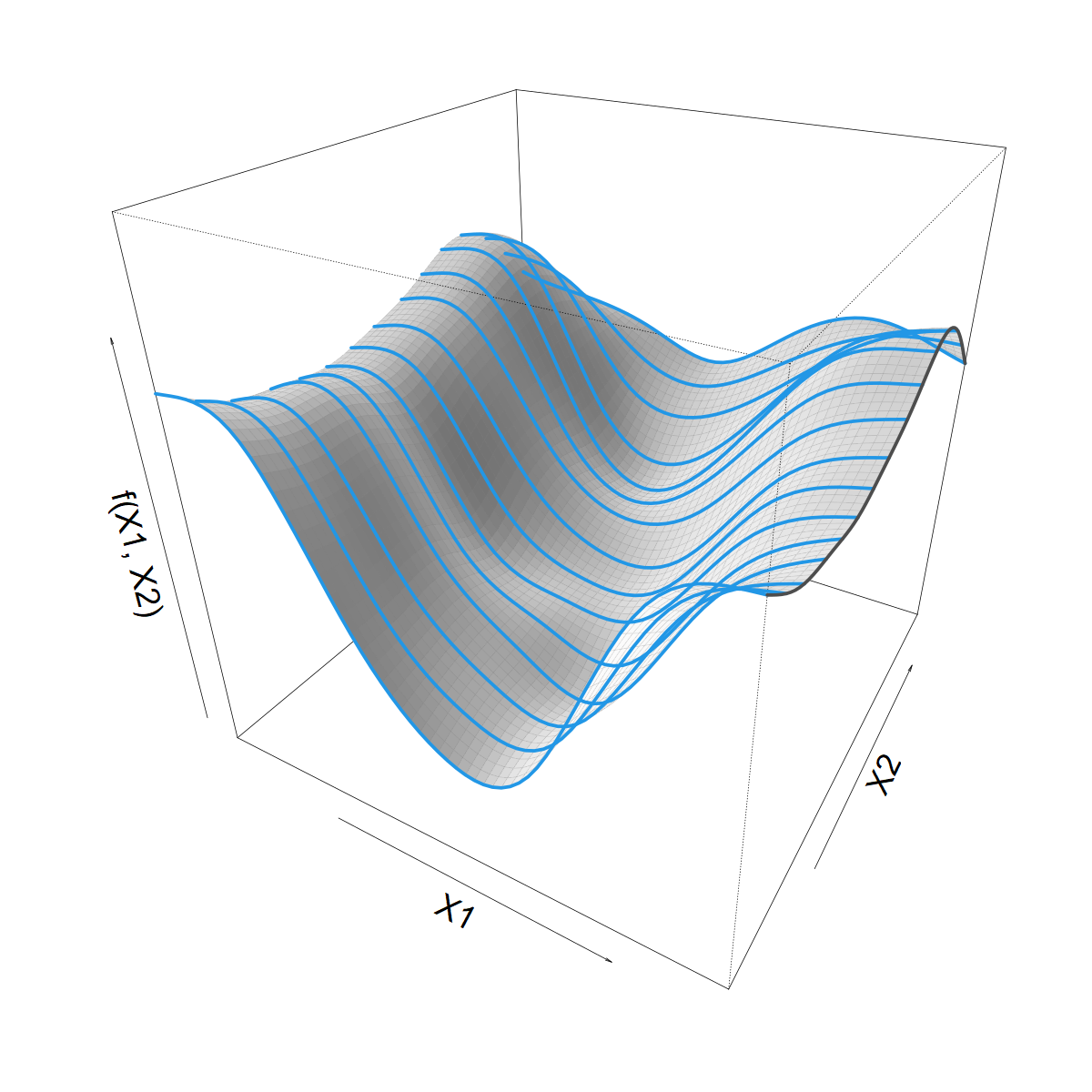
\[ s(X1) = \sum_k \beta_k b_k(X1) \]
\[ \beta_k(X2) = \sum_j \alpha_{jk} a_j(X2) \]
\[ f(X1,X2) = \]
\[ \sum_j \sum_k \alpha_{jk} a_j(X2) b_k(X1) \]
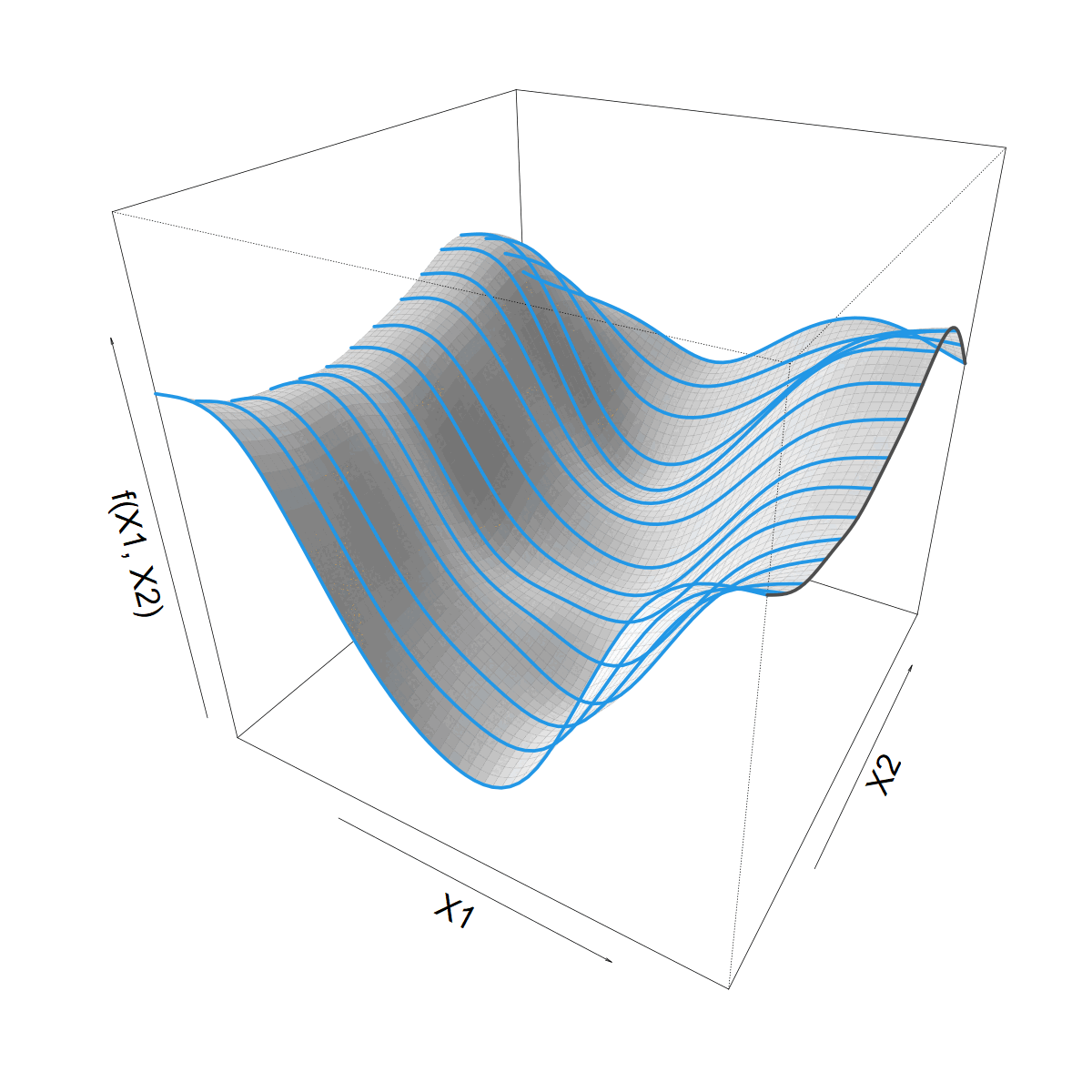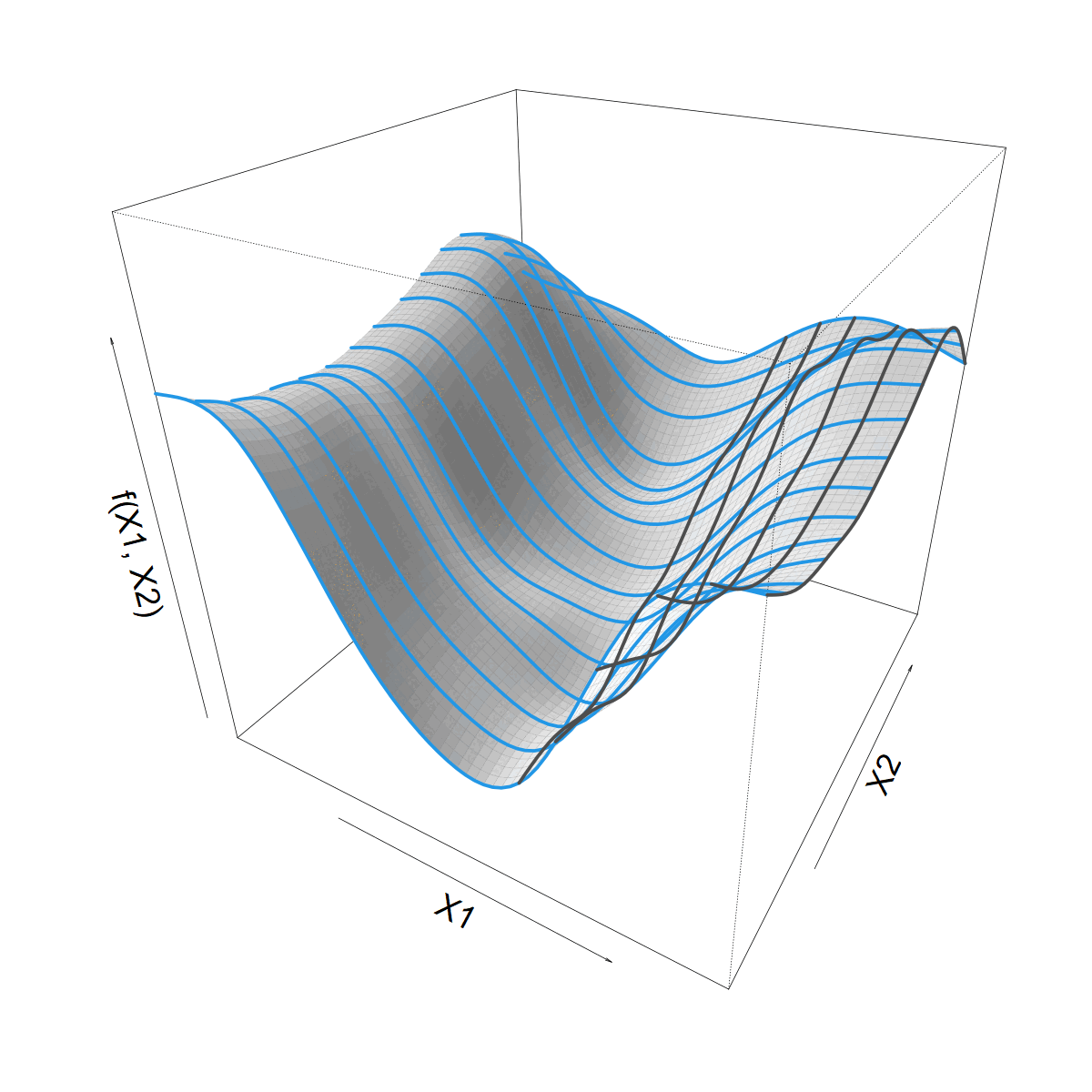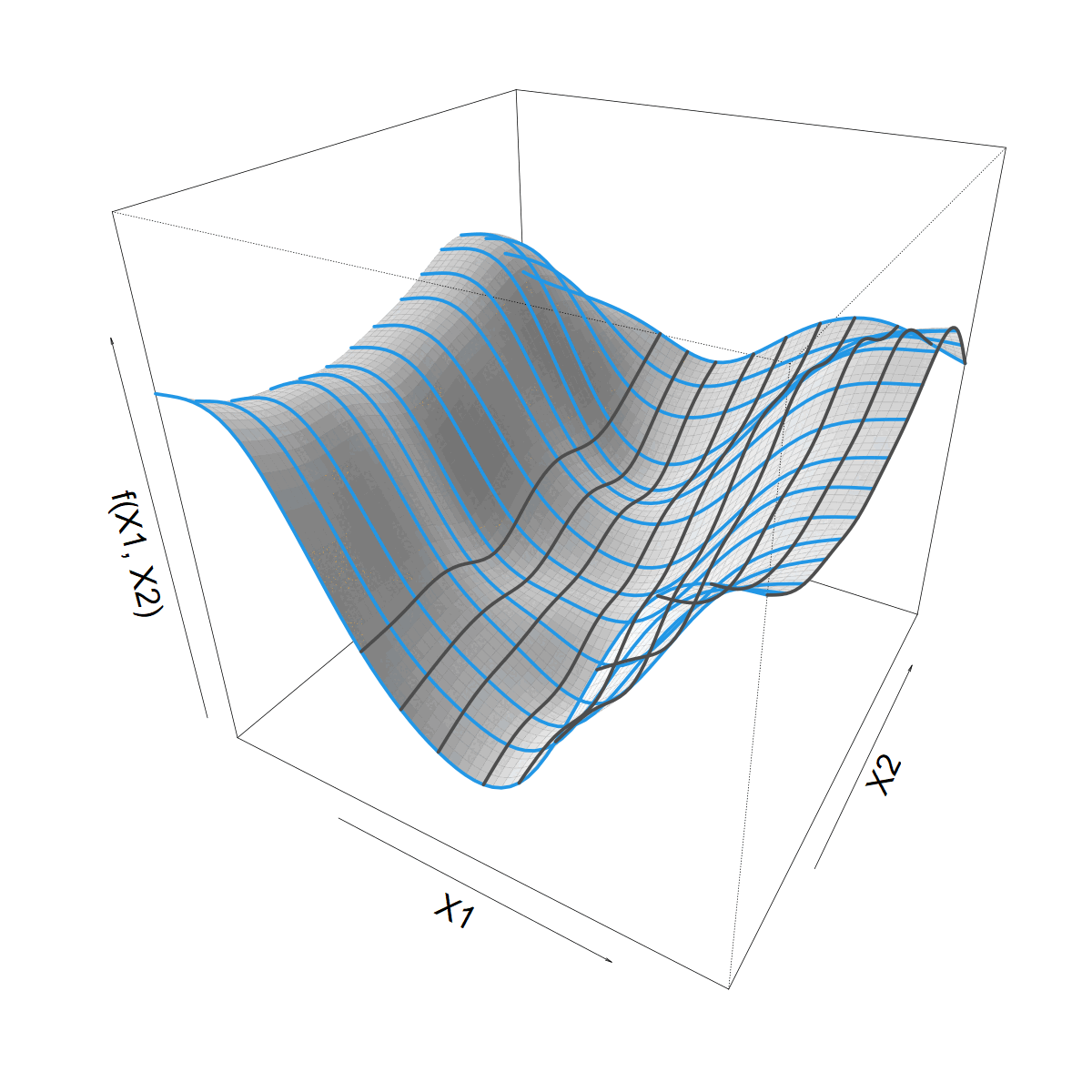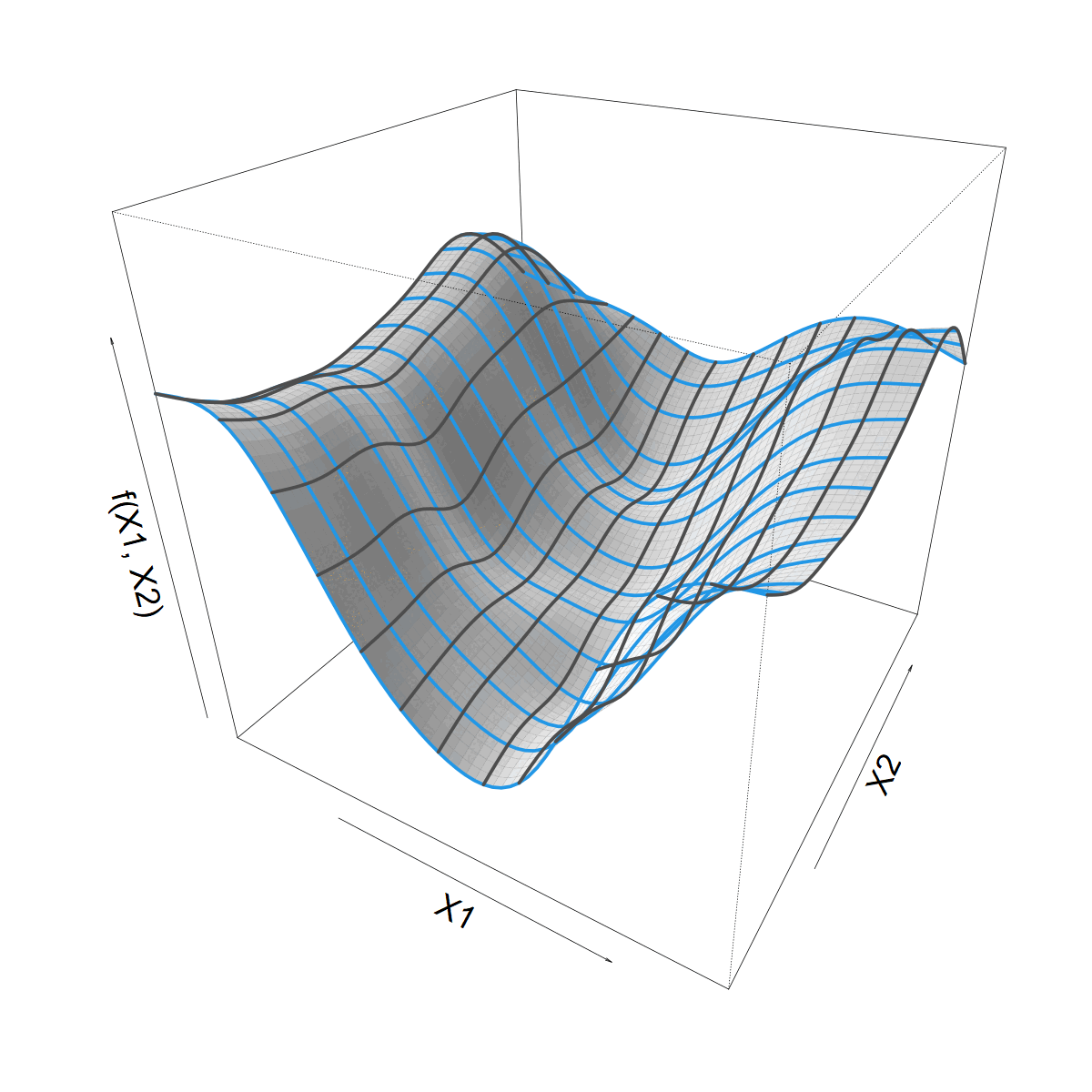
What about that Site bit?
- We can go from 2 to more dimensions
Siteis special because it’s a factor- but we can tensor splines and random effects
- Generates a copy of the smooth for each
Site- smoothing towards each other
- (see also Fergus’ talk!)
- This is a kind of hierarchical GAM
So, in R?
Setting-up the model in R

Setting-up the model in R
te(Dm, Tm, Sitem)would just be our 3D smoothDmis a distance matrixTmis a weekly matrixSitemis a site matrix
SSTmis a matrix of SSTs- Like before, but rows are different for each site
Recap
- We want 💅fancy💅 averages
- We can use any kind of smooth to make them
- We do need to faff making the lag matrices
by=just means “premultiply by”