lm(y ~ x1, data=dat)What are GAMs?
Biomathematics and Statistics Scotland
UK Centre for Ecology and Hydrology
Overview
- A very quick refresher on GLMs
- What is a GAM?
- How do GAMs work? (Roughly)
- What is smoothing?
- Fitting and plotting simple models
First…

A (very fast) refresher on (G)LMs
First, linear models
- Models that look like:
\[ y_i \sim \text{Normal}(\mu_i, \sigma_i) \]
- where:
\[ \mu_i = \beta_0 + \beta_1 x_{1i} + \beta_2 x_{2i} + \ldots \]
- Roughly: want to make \(y_i\) with weighted bits of the \(x_{ji}\)s
What is a Generalized Linear model (GLM)?
- Models that look like:
\[ y_i \sim \text{EF}(\mu_i, \sigma_i) \]
- where:
\[ g(\mu_i) = \beta_0 + \beta_1 x_{1i} + \beta_2 x_{2i} + \ldots \]
- \(g\) is a link function
- \(\text{EF}\) is an exponential family distribution
- Normal, Poisson, Binomial (all our usual friends)
What is a Generalized Linear model (GLM)?
- Models that look like:
\(y_i \sim \text{EF}(\mu_i, \sigma_i); \qquad g(\mu_i) = \beta_0 + \beta_1 x_{1i} + \beta_2 x_{2i} + \ldots\)
What does that mean?
The the response, \(\mu_i\), is assumed to be a linear combination (weighted by the \(\beta\)s) of the covariates, \(x_{ji}\), with an intercept (\(\beta_0\)).
Why bother with anything more complicated?
Is this linear?
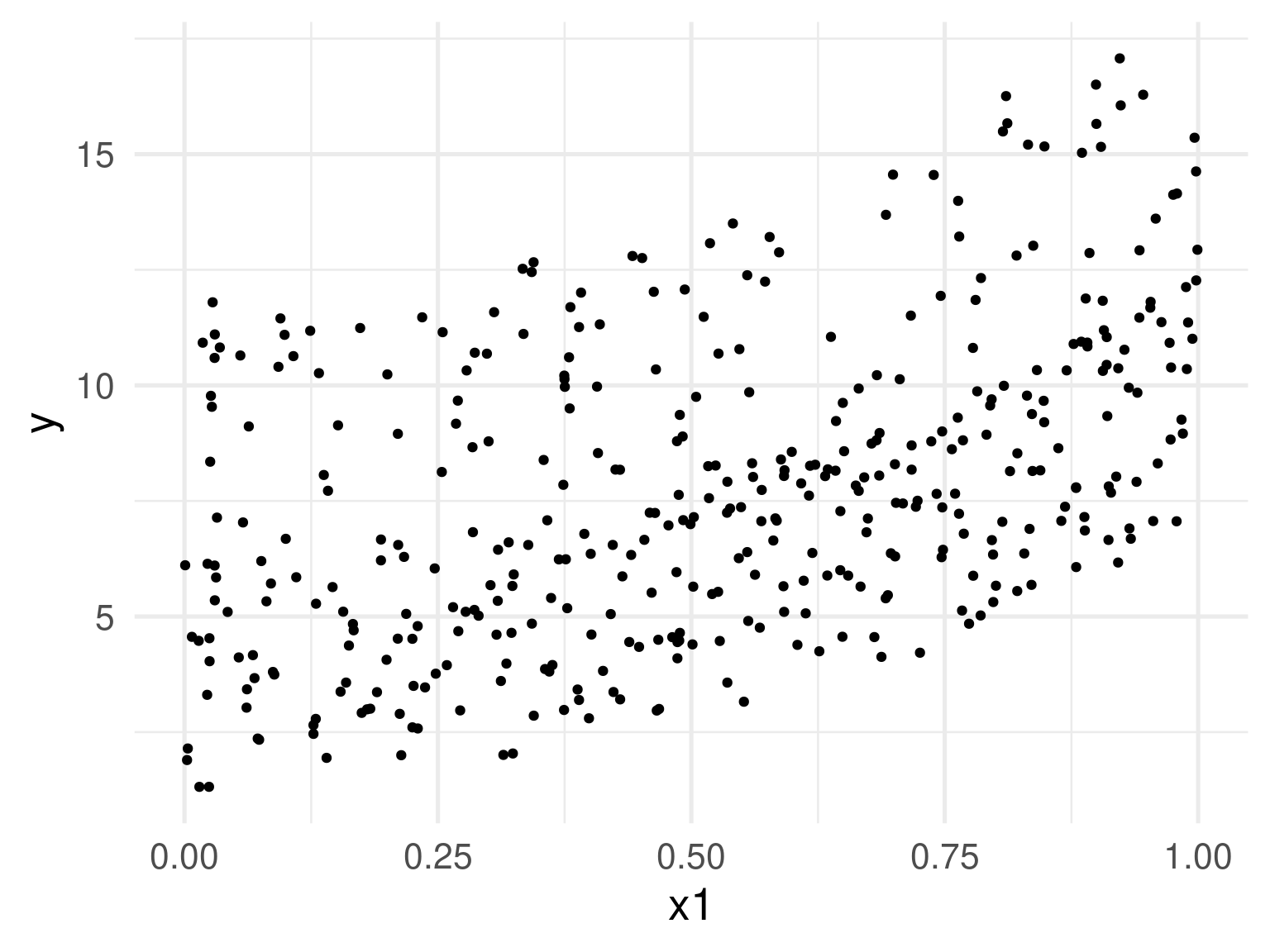
Is this linear? Maybe?
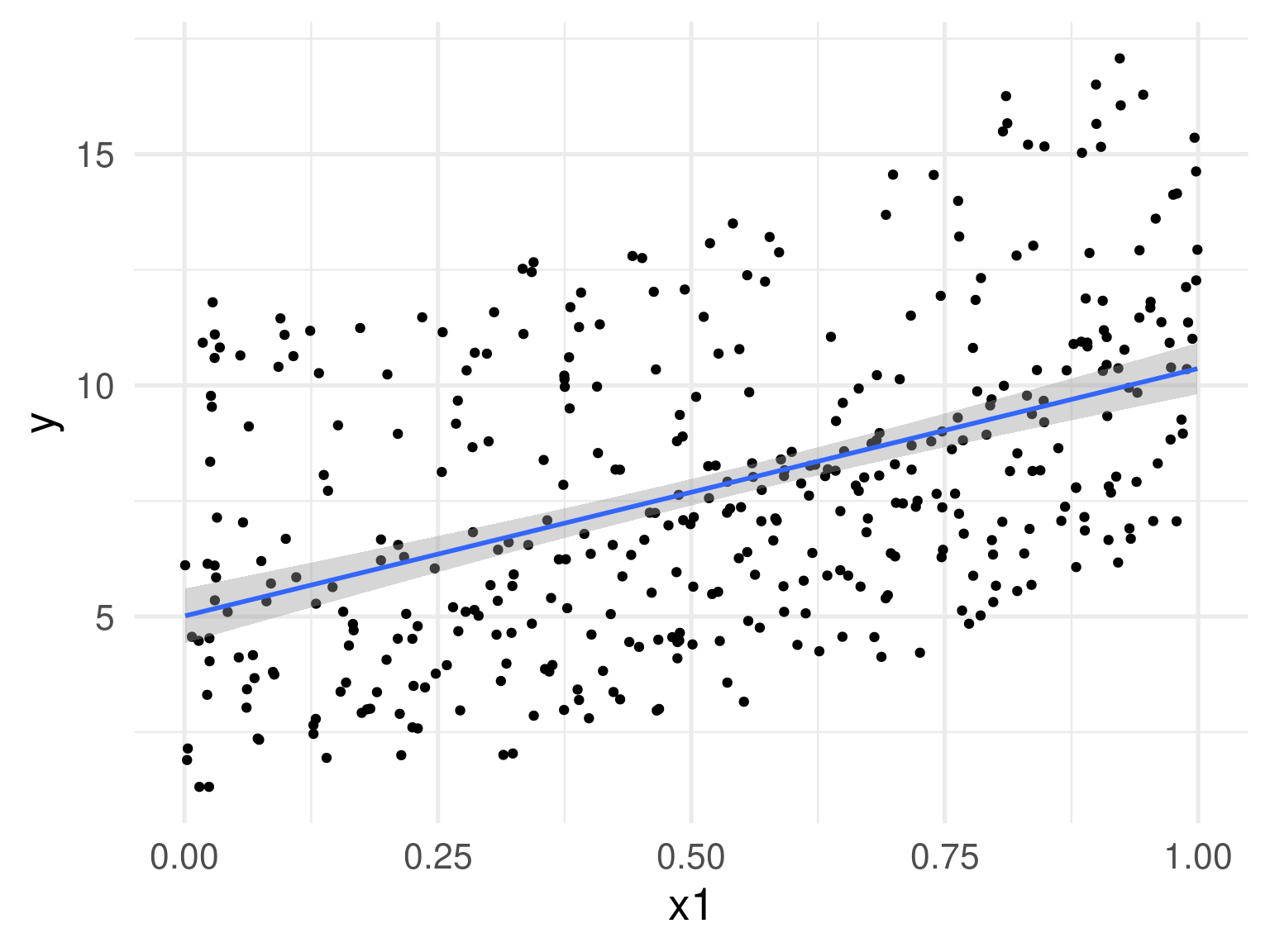
Is this linear? Maybe???
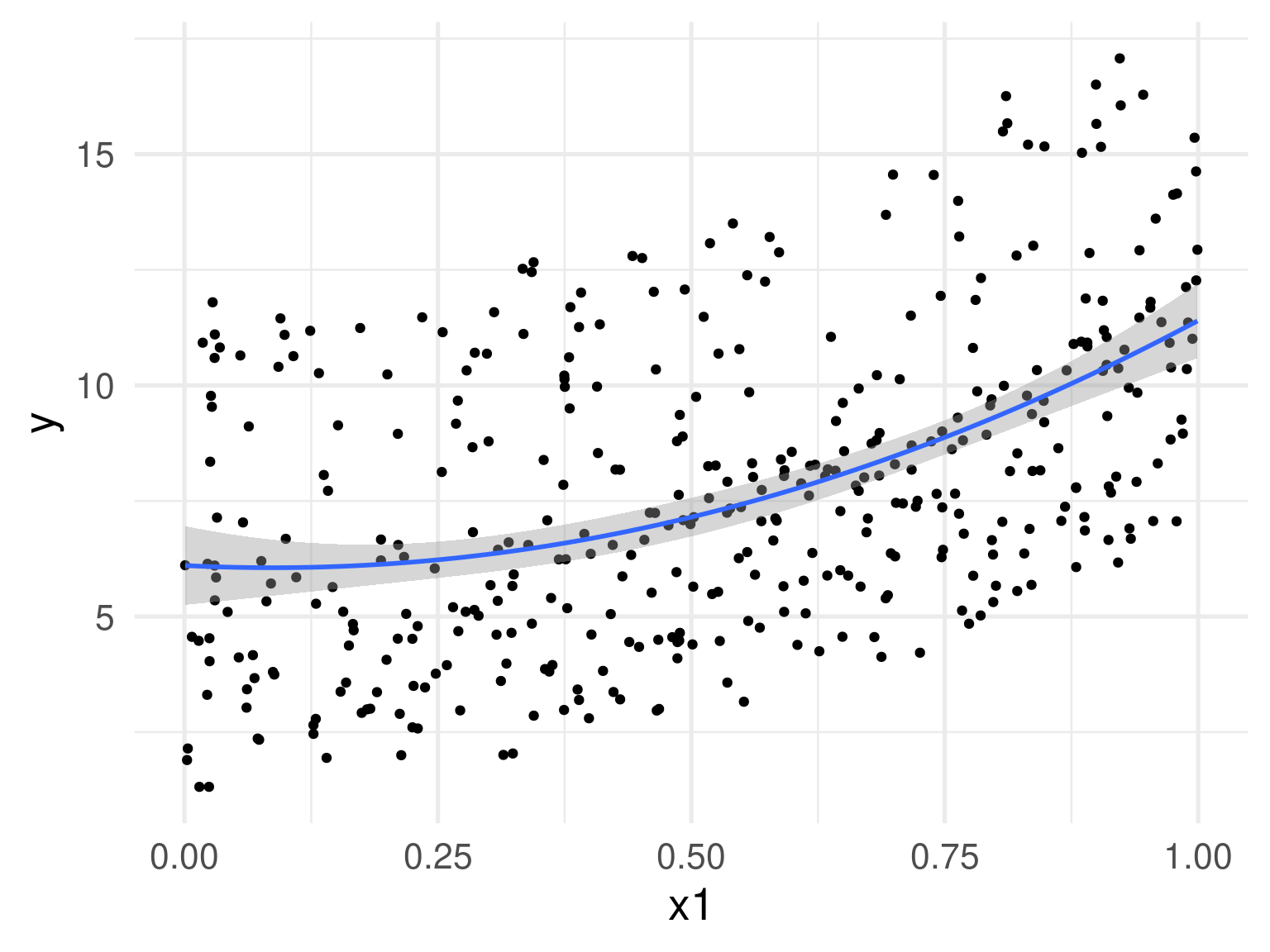
Is this linear? Maybe not.
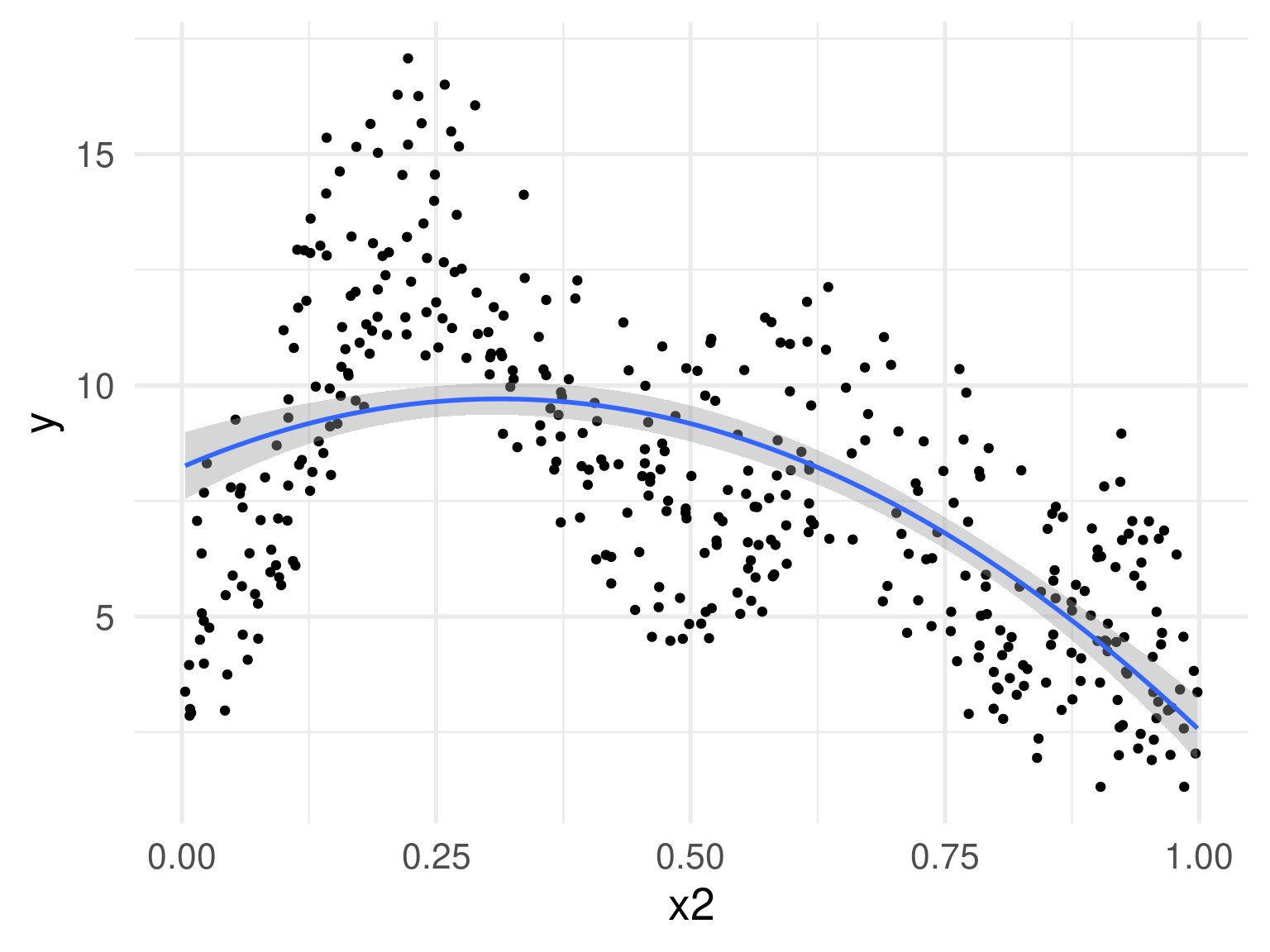
Can we keep adding polynomial terms?
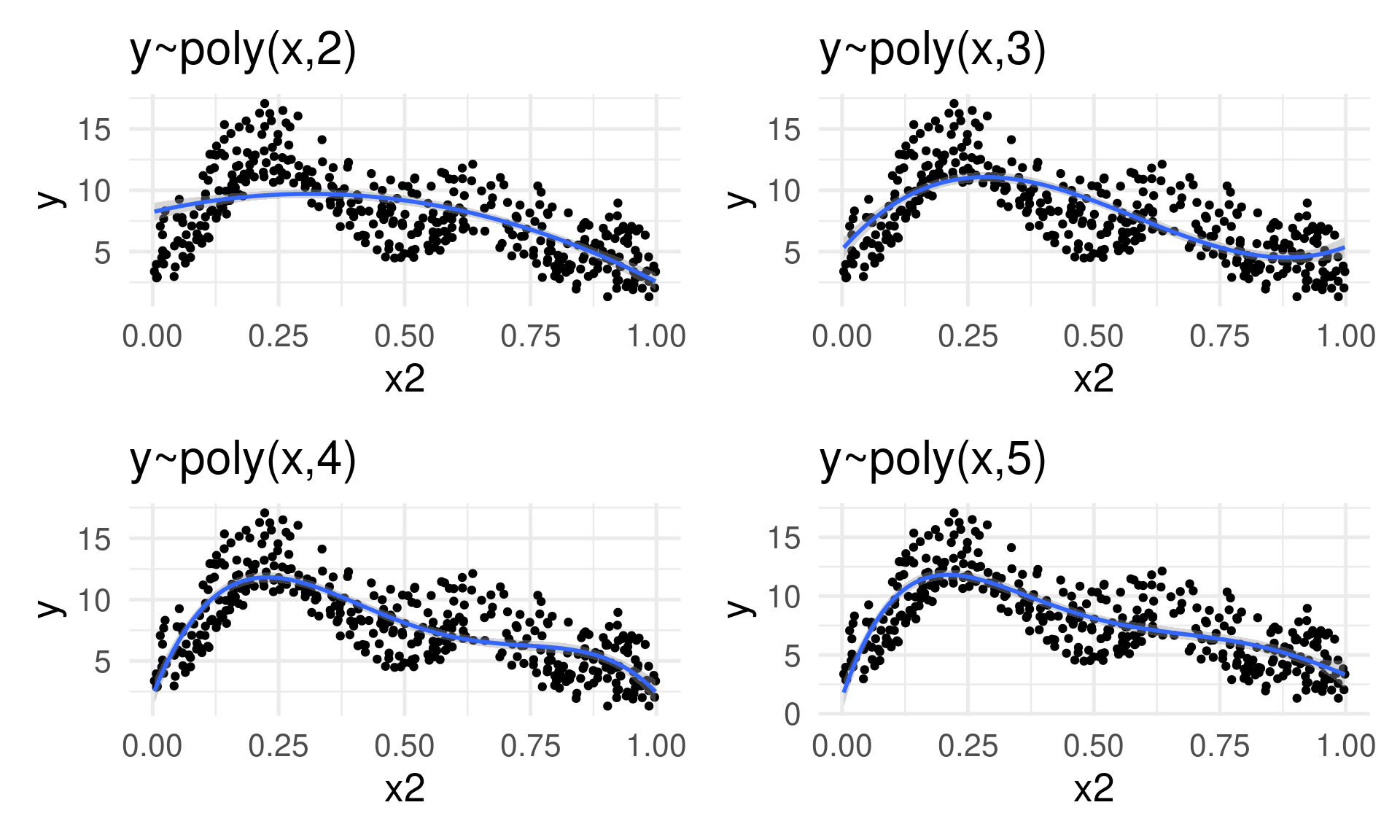
No
(for a few reasons)
Polynomial terms aren’t the answer
- No control of overfitting
- Computationally difficult to deal with (esp. with higher orders)
If we want to add wiggly effects, we use a GAM
Requirements
- Control for overfitting
- Flexible
- Fit within a framework we know
GAM examples
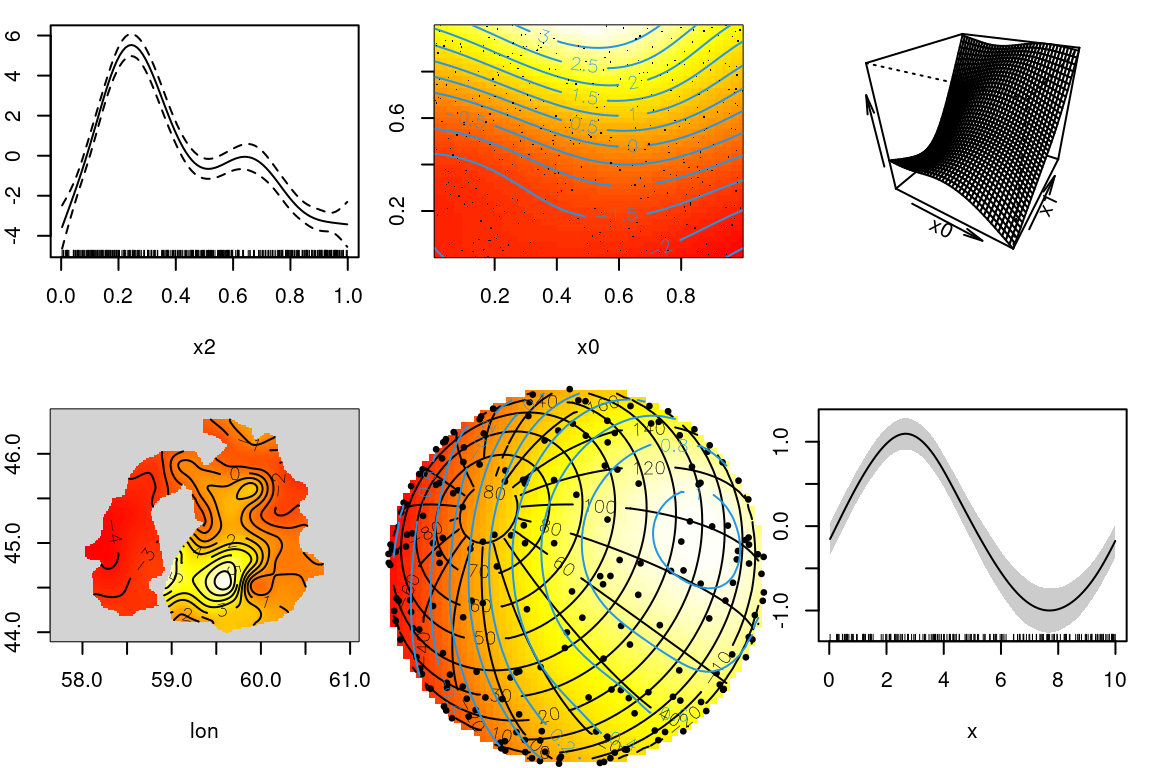

How do we make wiggly functions?
Splines (historical)
Splines (historical)

Photos from https://www.core77.com/posts/55368/When-Splines-Were-Physical-Objects
Smoothers
- Make flexible functions from adding simpler functions
\[ s(x) = \sum_k \beta_k b_k(x) \]
- Built from basis functions \(b_k(x)\)
- Only need to estimate \(\beta_k\)s
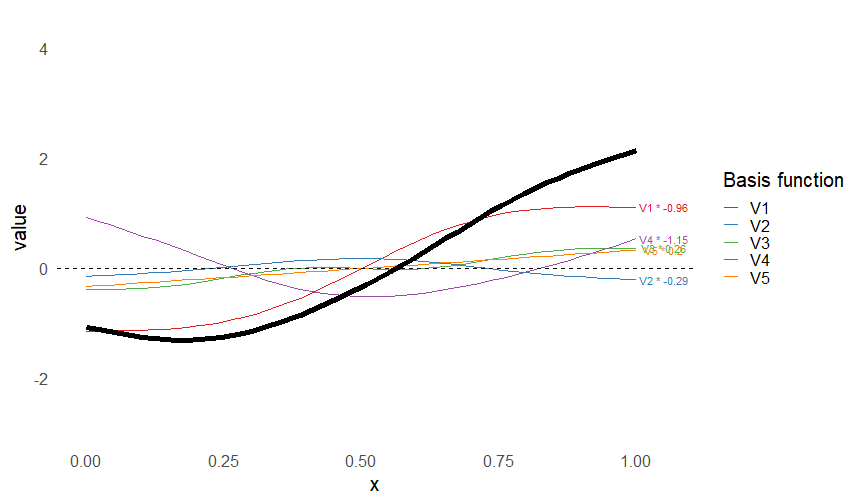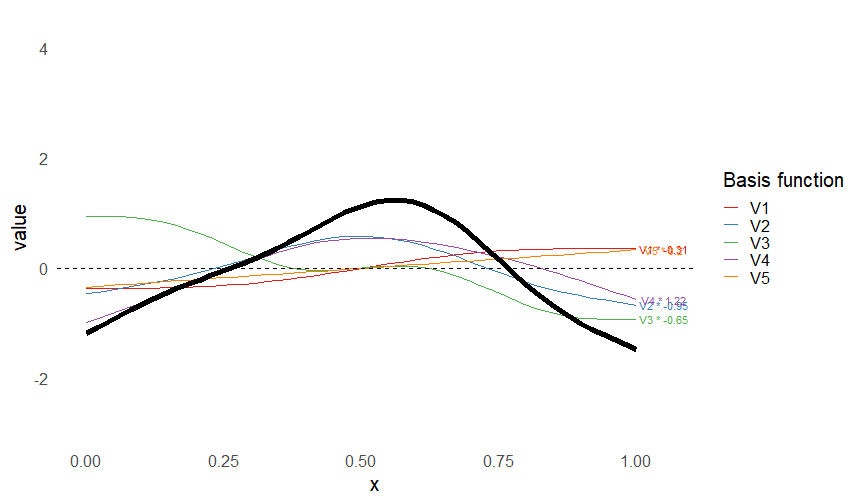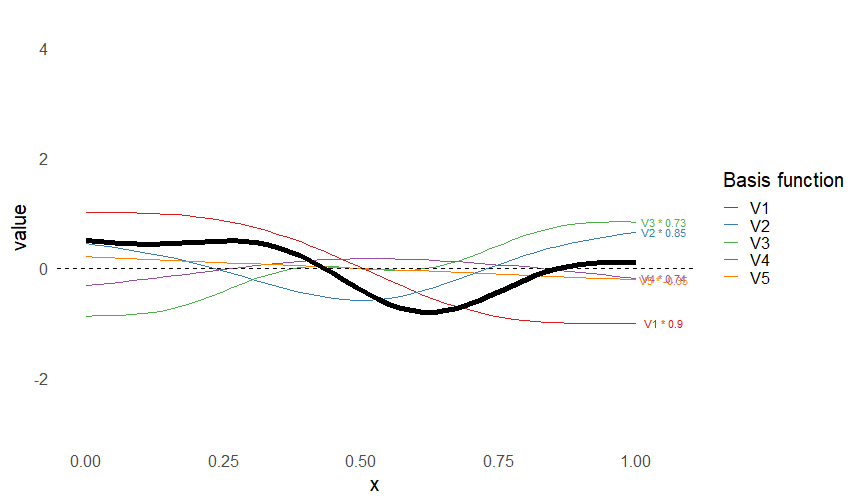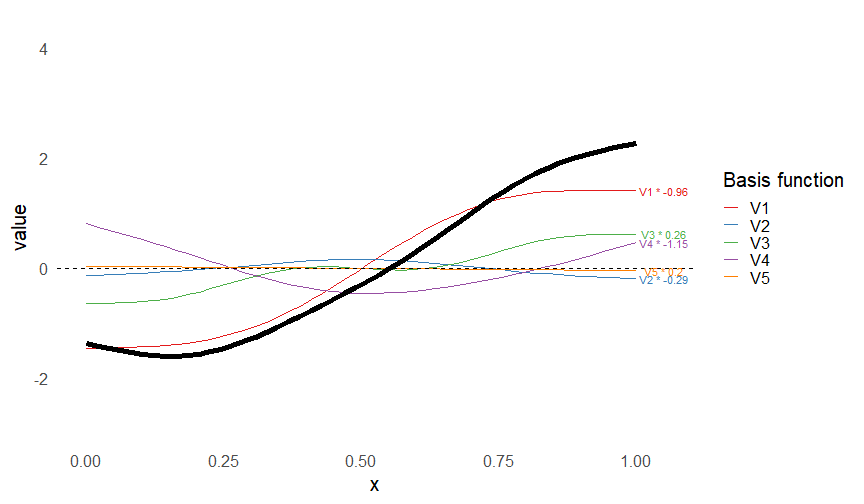
This leads to some very flexible functions!
Shiny app!
Basis-penalty smoothers
- How do we avoid overfitting?
- Measure wigglyness, then penalise!
\[ \lambda \int_\mathbb{R} \left( \frac{\partial^2 s(x)}{\partial x^2}\right)^2 \text{d}x\\ \]
where \(\lambda\) is a smoothing parameter
Basis-penalty smoothers
- How do we avoid overfitting?
- Measure wigglyness, then penalise!
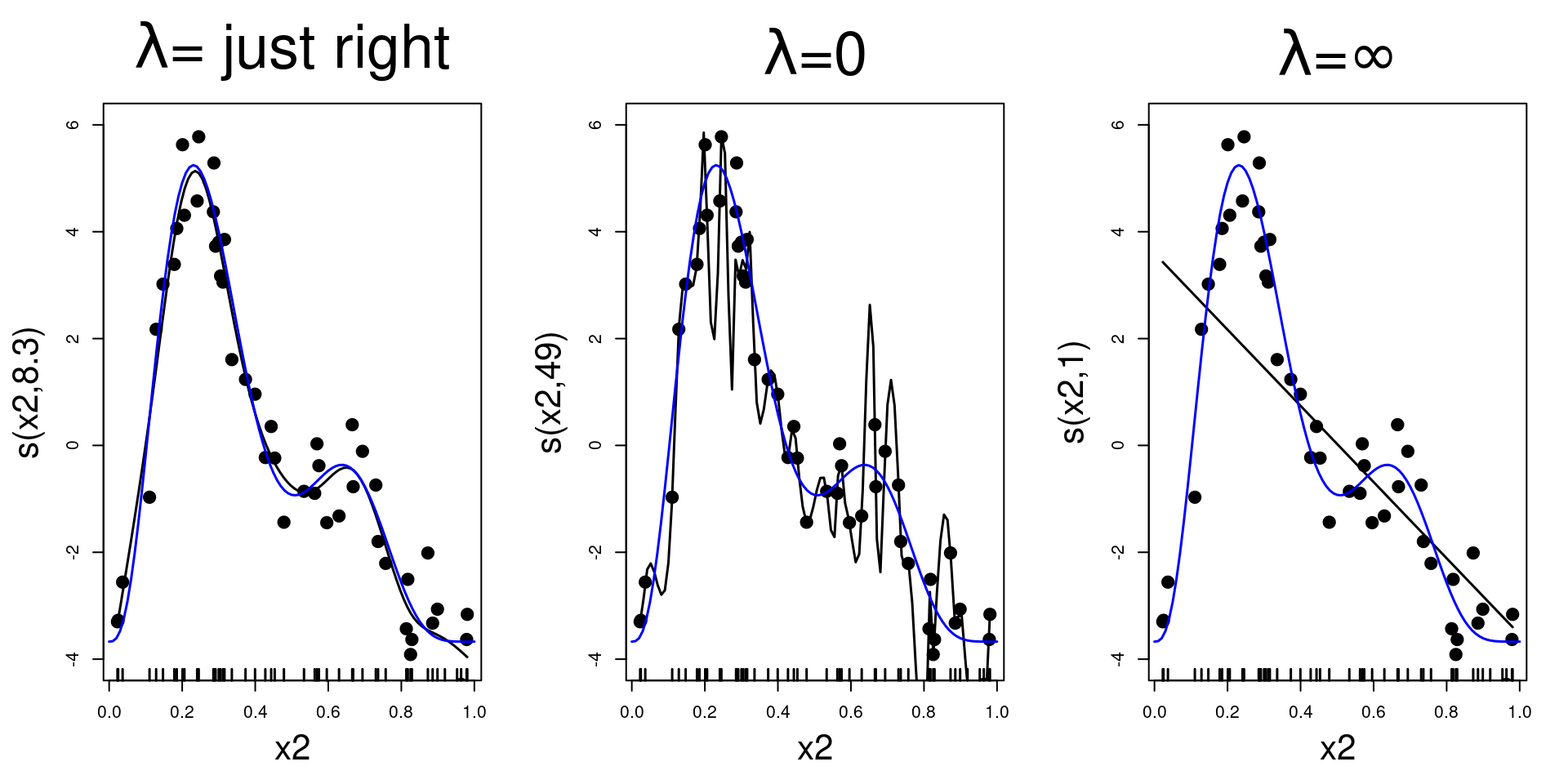
Effective degrees of freedom
- We need to set number of basis functions (\(k\))
- This sets the maximum complexity
- But we penalize so there is some shrinkage in the coefficients
- How many basis functions did we effectively use?
- effective degrees of freedom – EDF
Effective degrees of freedom
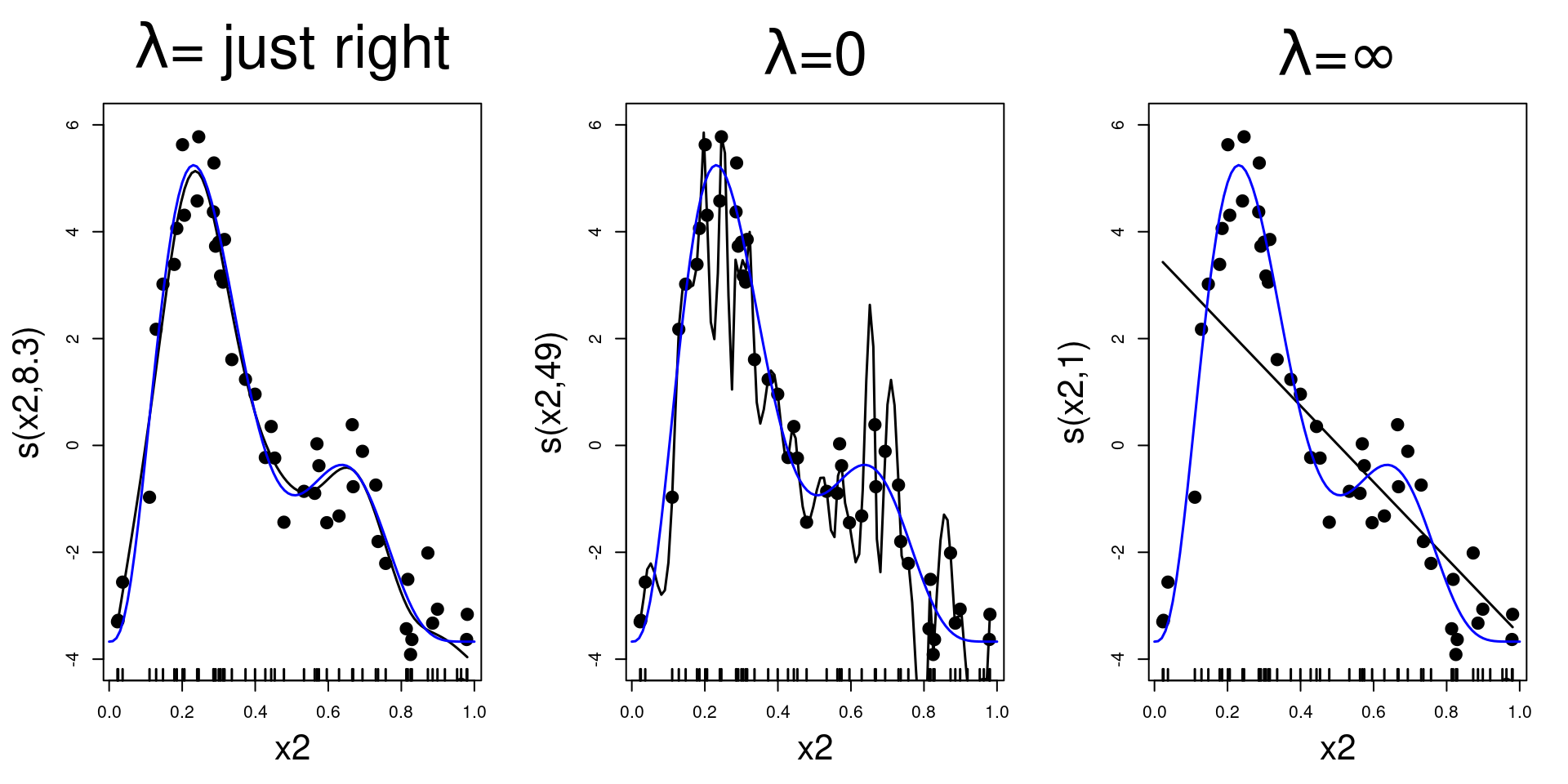
Putting that in a model
\[ \mathbb{E}(Y_i) = g^{-1}\left[\beta_0 + \sum_j s_j(x_{ij})\right] \]
- \(Y_i \sim\) some (extended?) exponential family
- \(g\) is a link function
- \(\beta_0\) is an intercept
- \(s_j(x_{ij})\) are smooths!
Okay, what about practical stuff…
Fitting GAMs in R
- Lots of packages exist:
gam,mgcv,R-INLA,brms,gamlss - Here we’ll focus on
mgcv- R package, by Simon Wood (Edinburgh)
- Uses familiar
glm()-type syntax Recommended(ships with R)- Very fast (and parallelisable)
The data we’re going to look at
UKCEH data: COSMOS-UK https://cosmos.ceh.ac.uk/

Dataset from: https://catalogue.ceh.ac.uk/documents/5060cc27-0b5b-471b-86eb-71f96da0c80f
COSMOS-UK
Using ✨cosmic rays✨ to measure moisture
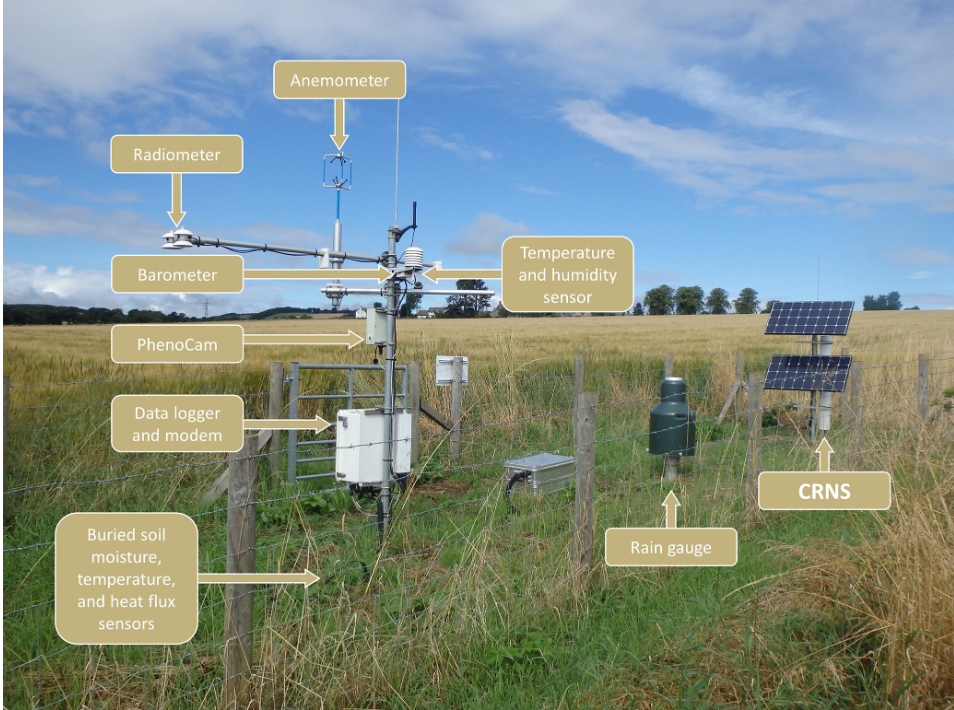
COSMOS-UK data
# A tibble: 127,228 × 16
SITE_ID SITE_NAME LONGITUDE LATITUDE NORTHING EASTING ALTITUDE LAND_COVER
<fct> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <fct>
1 ALIC1 Alice Holt -0.858 51.2 139985 479950 80 Broadleaf woo…
2 ALIC1 Alice Holt -0.858 51.2 139985 479950 80 Broadleaf woo…
3 ALIC1 Alice Holt -0.858 51.2 139985 479950 80 Broadleaf woo…
4 ALIC1 Alice Holt -0.858 51.2 139985 479950 80 Broadleaf woo…
5 ALIC1 Alice Holt -0.858 51.2 139985 479950 80 Broadleaf woo…
6 ALIC1 Alice Holt -0.858 51.2 139985 479950 80 Broadleaf woo…
7 ALIC1 Alice Holt -0.858 51.2 139985 479950 80 Broadleaf woo…
8 ALIC1 Alice Holt -0.858 51.2 139985 479950 80 Broadleaf woo…
# ℹ 127,220 more rows
# ℹ 8 more variables: SOIL_TYPE <fct>, DATE_TIME <date>, PRECIP <dbl>,
# SNOW_DEPTH <dbl>, COSMOS_VWC <dbl>, ndate <dbl>, month <dbl>, year <dbl>COSMOS-UK data
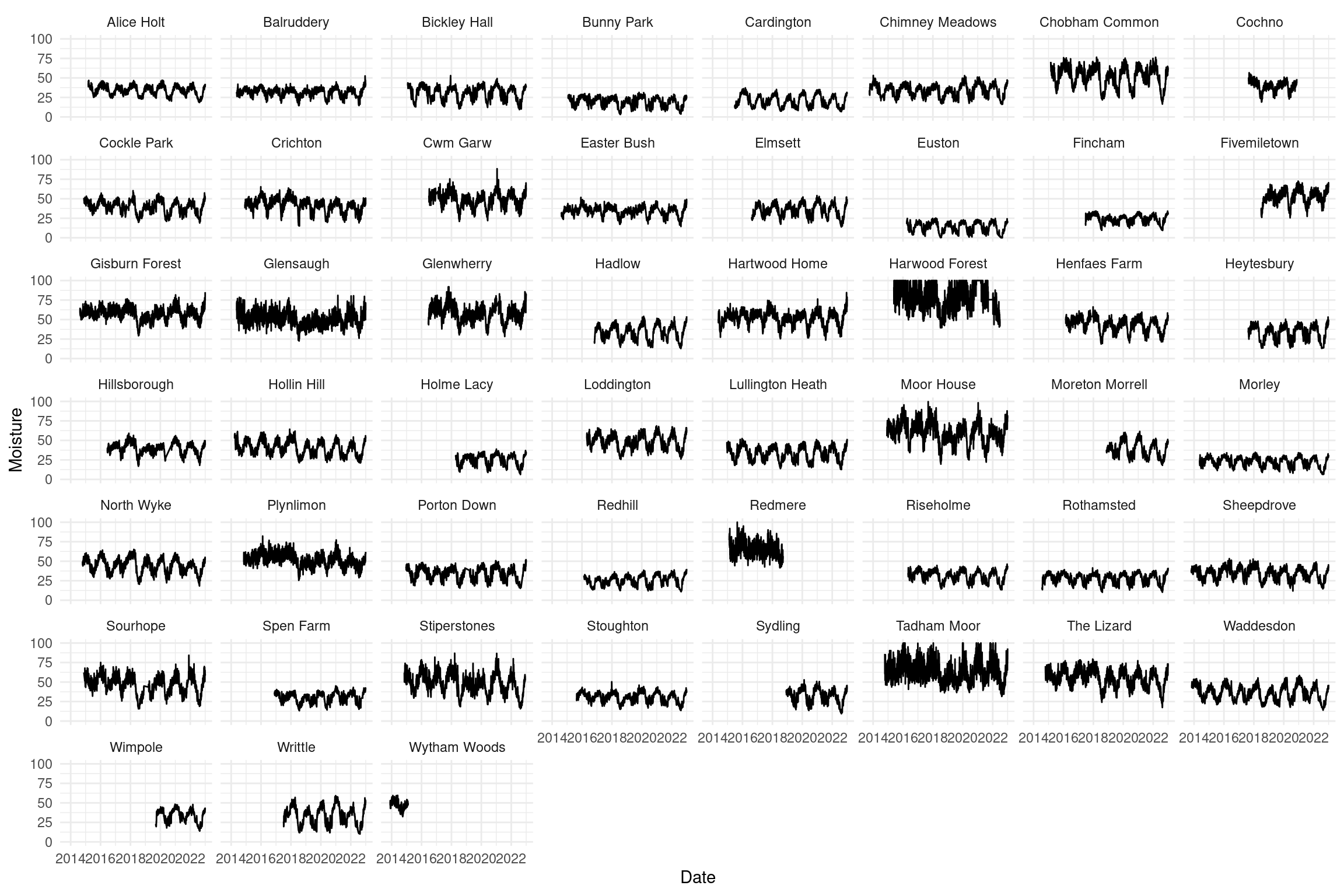
Let’s try to fit a model to the soil data
- Simple: predict moisture as a function of day (for one site)
- Ignore some data structure (for now)
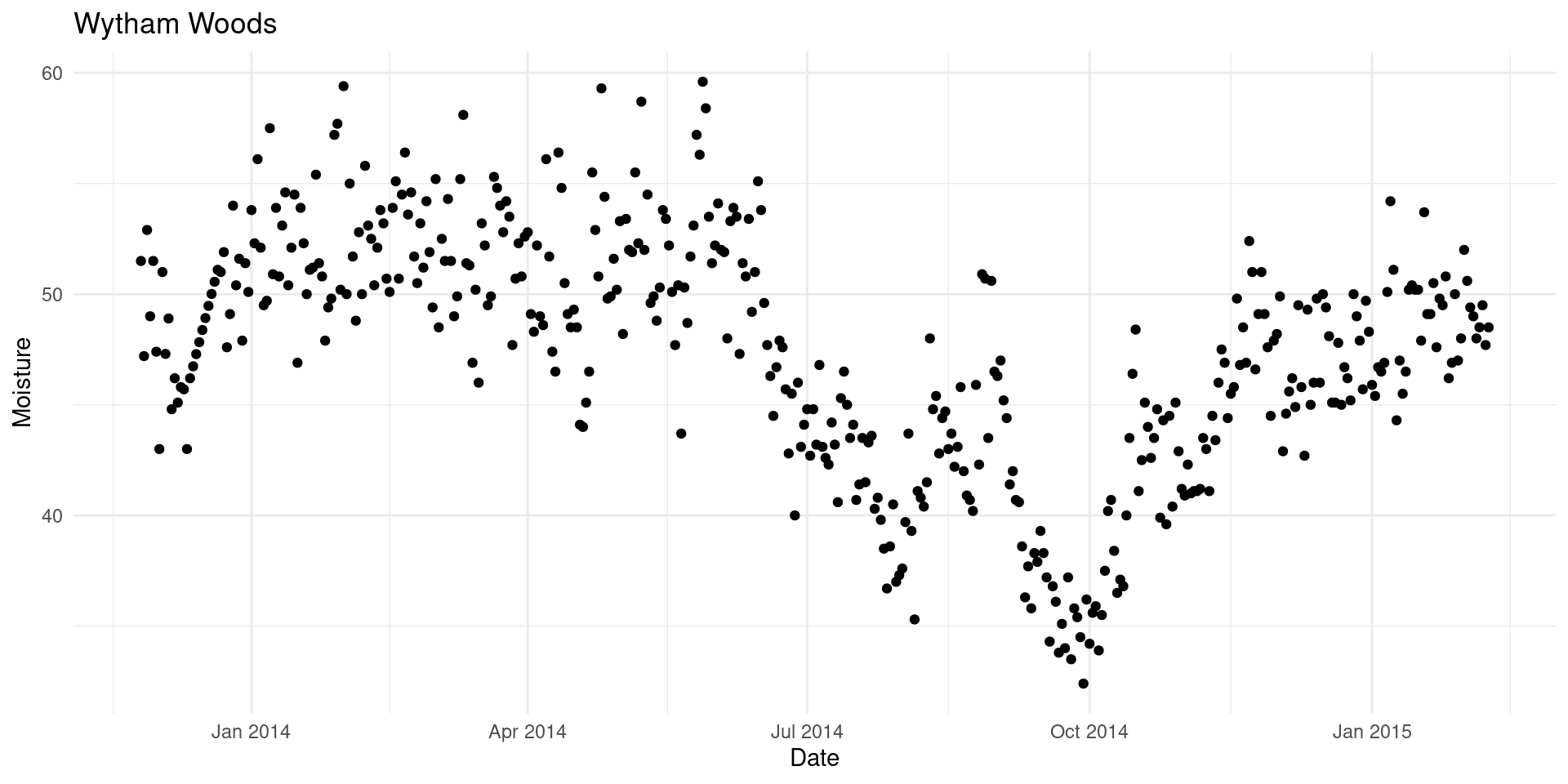
The model
- Denote:
- day by \(t\)
- moisture on day \(t\) is \(m_{t}\)
- Assume \(m_{t} \sim N(\mu_t, \sigma)\)
- Smooth effect of day: \(s(t)\)
The model
- Denote:
- day by \(t\)
- moisture on day \(t\) is \(m_{t}\)
- Assume \(m_{t} \sim N(\mu_t, \sigma)\)
- Smooth effect of day: \(s(t)\)
Our model is then: \[ \mathbb{E}(\mu_t) = \beta_0 + s(t) \]
Fitting a model with gam
- If we know how to use
glm, we know how to usegam - We need a
formulaand somedata - We can denote smooths with
s()- e.g.,
s(x)
- e.g.,
What did that do?
Family: gaussian
Link function: identity
Formula:
COSMOS_VWC ~ s(ndate)
Parametric coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 47.3525 0.1524 310.7 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Approximate significance of smooth terms:
edf Ref.df F p-value
s(ndate) 8.167 8.812 95.1 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
R-sq.(adj) = 0.656 Deviance explained = 66.3%
-REML = 1150.5 Scale est. = 10.218 n = 440What did that do?
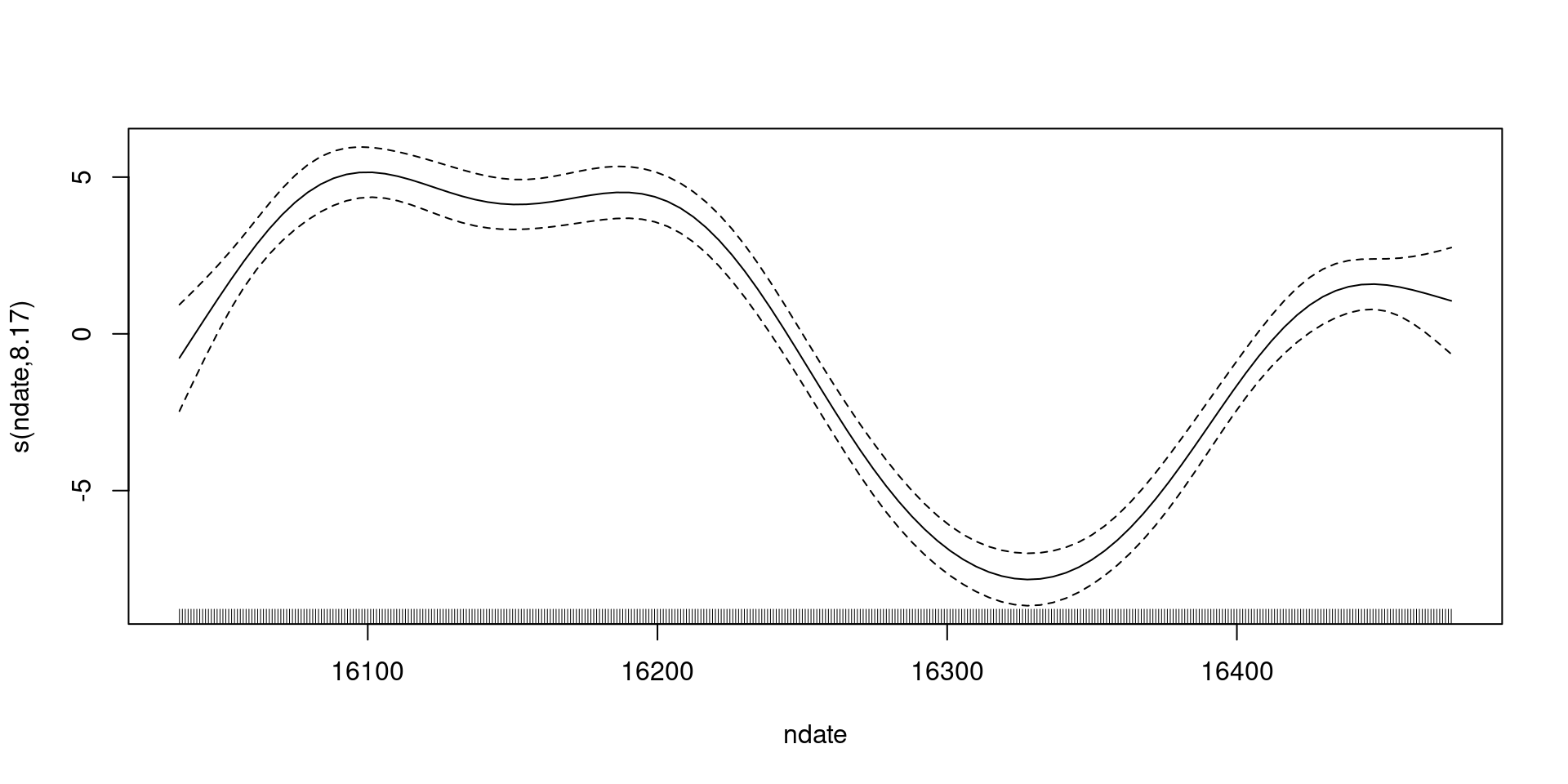
What did that do?
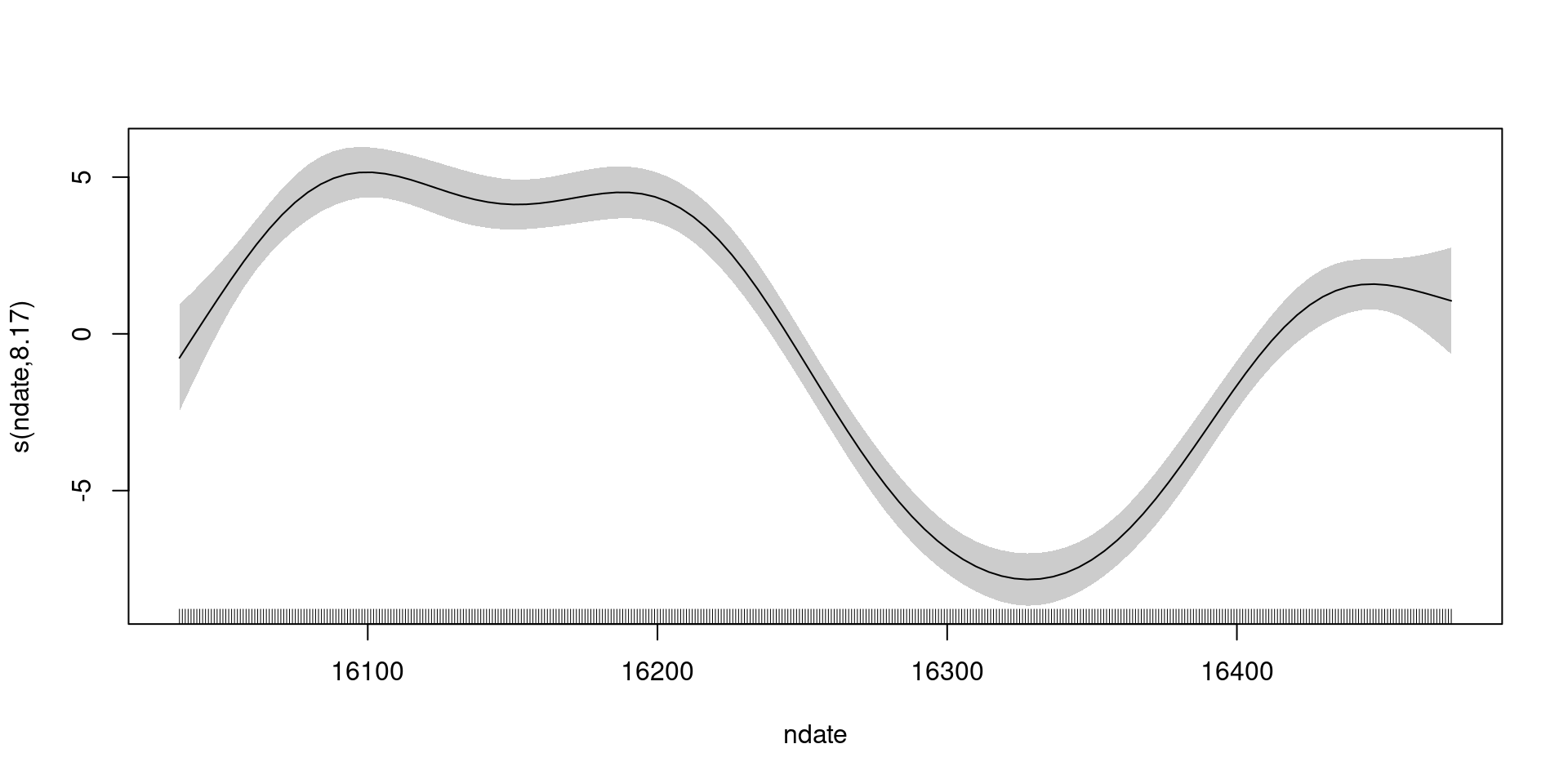
Was the smooth wiggly enough?
Method: REML Optimizer: outer newton
full convergence after 7 iterations.
Gradient range [-3.367285e-07,8.300716e-08]
(score 1150.533 & scale 10.2183).
Hessian positive definite, eigenvalue range [3.13693,219.0595].
Model rank = 10 / 10
Basis dimension (k) checking results. Low p-value (k-index<1) may
indicate that k is too low, especially if edf is close to k'.
k' edf k-index p-value
s(ndate) 9.00 8.17 0.5 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Let’s double k
Family: gaussian
Link function: identity
Formula:
COSMOS_VWC ~ s(ndate, k = 20)
Parametric coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 47.3525 0.1289 367.4 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Approximate significance of smooth terms:
edf Ref.df F p-value
s(ndate) 17.43 18.68 71.43 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
R-sq.(adj) = 0.754 Deviance explained = 76.4%
-REML = 1099.9 Scale est. = 7.3089 n = 440What do our models look like?
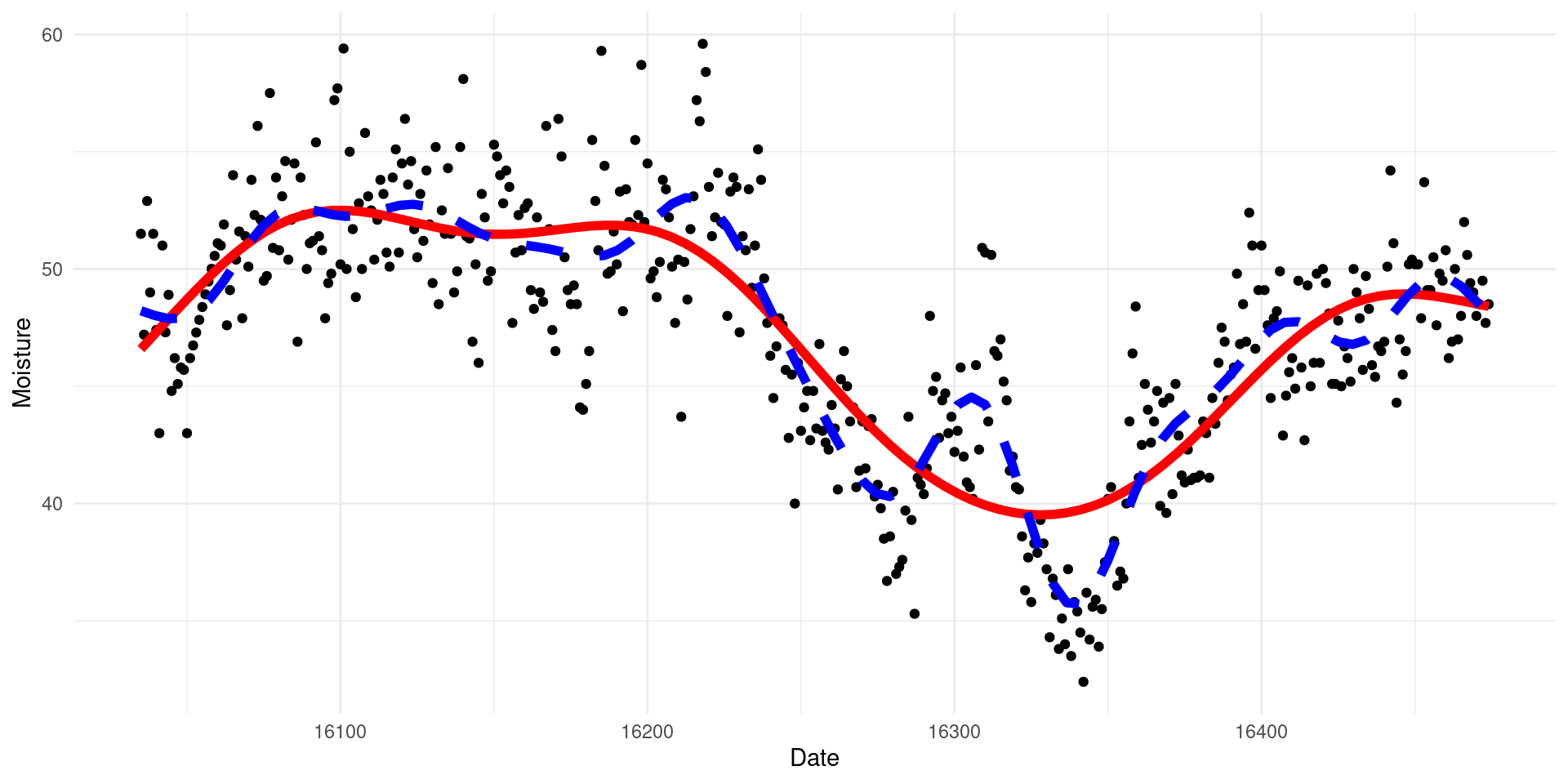
(you’ll learn how to do this later!)
Recap
- GAMs are GLMs with extra wiggles
- We use basis functions to model flexible effects
- We need to penalize our fit to avoid overfitting
gam()fits a GAMsummary()gives us information about the fitplot()makes basic plots